Complex I
Complex I catalyzes the oxidation of NADH and the reduction of quinone coupled to charge translocation across the membrane. Deficiencies on this complex have been shown to be implicated in several pathologies, namely neurodegenerative diseases such as Leber’s hereditary optic neuropathy, Parkinson and Dystonia disorders. The study of this enzyme is currently a hot-topic in the bioenergetics’ field, with a recent gathering of structural and functional data, including its X-ray structure.
We have been studying complex I using Rhodothermus marinus as a model system, namely investigating its electron transfer kinetics and quinone reduction, H+ and Na+ translocation and the coupling mechanism of electron transfer to ion transport. We have recently extended our approach to complex I from Escherichia coli and Paracoccus denitrificans, the most studied prokaryotic complexes I.
Complex I is an energy transducting enzyme present in the three domains of life. This enzyme catalyzes the oxidation of NADH and the reduction of quinone coupled to charge translocation across the membrane. In this way, it contributes to the establishment of the transmembrane difference of electrochemical potential which is used for ATP synthesis, solute transport and motility.
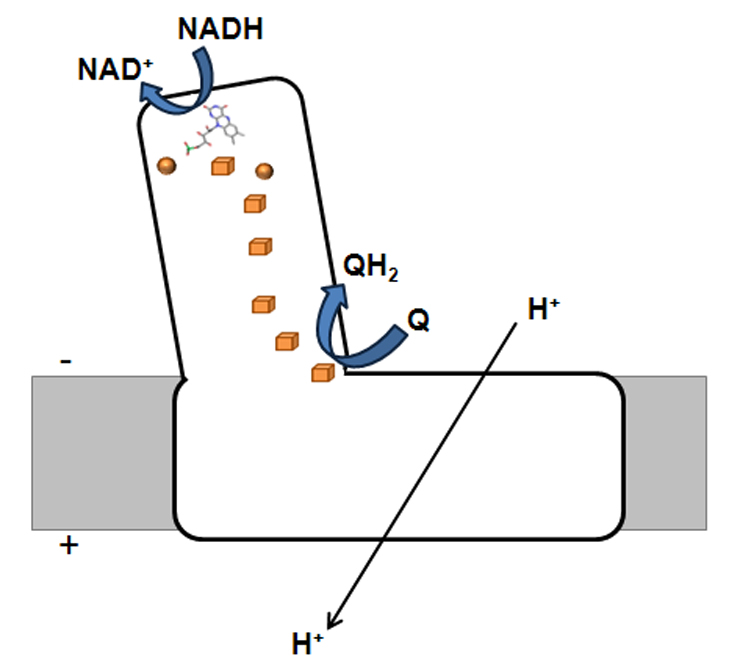
Figure 1 - Schematic representation of bacterial complex I. The orange spheres and cubes represents [2Fe-2S]2+/1+ and [4Fe-4S]2+/1+, respectively.
This respiratory complex is an L-shaped membrane enzyme, consisting of a peripheral and a membrane arm (Figure 1). The peripheral arm contains a series of iron-sulphur centres (binuclear and tetranuclear ones, named N1a, N1b, N2, N3, N4, N5, N6a and N6b) and a FMN, at the catalytic site where NADH is oxidized. The membrane arm includes the charge translocating machinery. The bacterial complex I is, generally, composed of 14 subunits (named Nqo1-14 or NuoA-N), which may be considered modular units and which are also present in other complexes. Nqo1, 2 and 3 constitute the electron input module and with the exception of the C-terminal part of Nqo3, are homologous to the subunits of soluble NAD+ reducing hydrogenases. The C-terminal part of Nqo3 is homologous to molybdopterin enzymes. Nqo4, 5, 6, 8 and 9 are homologous to subunits of the membrane-bound NiFe hydrogenases, whereas the subunits Nqo11, 12, 13 and 14 are homologous to Mrp-like Na+/H+ antiporters. No homologues have been observed for subunits Nqo7 and Nqo10.
The nature of the ion(s) that is translocated by this enzyme is still a highly discussed issue, being protons and sodium ions possible candidates. It has long been established that complex I translocates H+, but the H+/electron stoichiometry has only been determined for the mitochondrial system. Few years ago the idea that all complexes I from all organisms only translocate H+ was challenged, by the observation that complex I from Klebsiella pneumoniae may translocate Na+ instead, being this the coupling ion of the system.
We have recently shown that, in fact, some bacterial complexes I are capable of H+ and Na+ translocation, but to opposite directions, being the H+ the coupling ion (Figure 2). We developed an original approach using 23Na-NMR spectroscopy, which was the first reported use of this technique to monitor substrate-driven Na+ transport by membrane vesicles. We observed that Rhodothermus marinus complex I has two H+ translocating sites, one operating independently of the presence of Na+ and the other working as a Na+/H+ antiporter. In order to establish whether the antiporter site was exclusive of R. marinus complex I, we addressed ion translocation by the two most studied bacterial complexes I. We observed that complex I from Escherichia coli also presents the antiporter activity, but that from Paracoccus denitrificans does not. We hypothesized a correlation between the type of quinone used as substrate and the presence of the antiporter activity (Figure 2). Furthermore, from our results using a typical inhibitor of Na+/H+ antiporters we hypothesized that energy coupling in complex I occurs through an indirect mechanism.
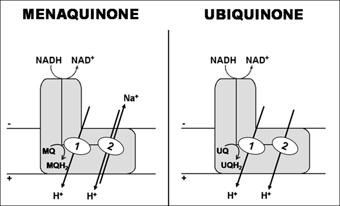
Figure 2 - Schematic representation of possible mechanisms for the transducing process of complex I, considering menaquinone or ubiquinone as the electron acceptor (R. marinus and E. coli complexes I use menaquinone as electron acceptor while P. denitrificans complex I use ubiquinone). Two proton translocation sites: 1- Independent of the presence of Na+; 2 - Working as a Na+/H+ antiporter.



