Research
Publications
You are invited to browse Manuel Melo's Google Scholar profile. Google does a much better job of keeping track of publications, and citations thereto, than we would ever be able to!
The Martini model
The Martini force field is a modern and powerful coarse-grain MD model. It easily permits the simulation of events well into the hundreds of microseconds.
This performance, typically 100 to 1000-fold faster than atomistic-detail models, is crucial to the type of problems we're interested in.
Speedup is achieved by clustering together groups of (typically 4) atoms as single beads. By throwing away some of the finer detail in a judicious manner relevant physical behavior is retained, at much lower computation cost.
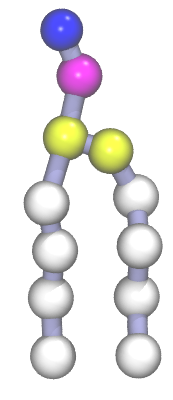
A Martini DPPC phospholipid
Antimicrobial peptides
This class of antibiotic peptides, amphipathic and highly cationic, has a peculiarly high affinity for bacterial membranes. It is accepted than in most cases membrane disruption is the main antimicrobial mechanism.
However, very little is known about the precise molecular-level organization of these peptides when pores are formed, or the bilayer is otherwise destroyed. Several models have been proposed; MD simulations have provided some of the most detailed accounts to date of antimicrobial peptide action.
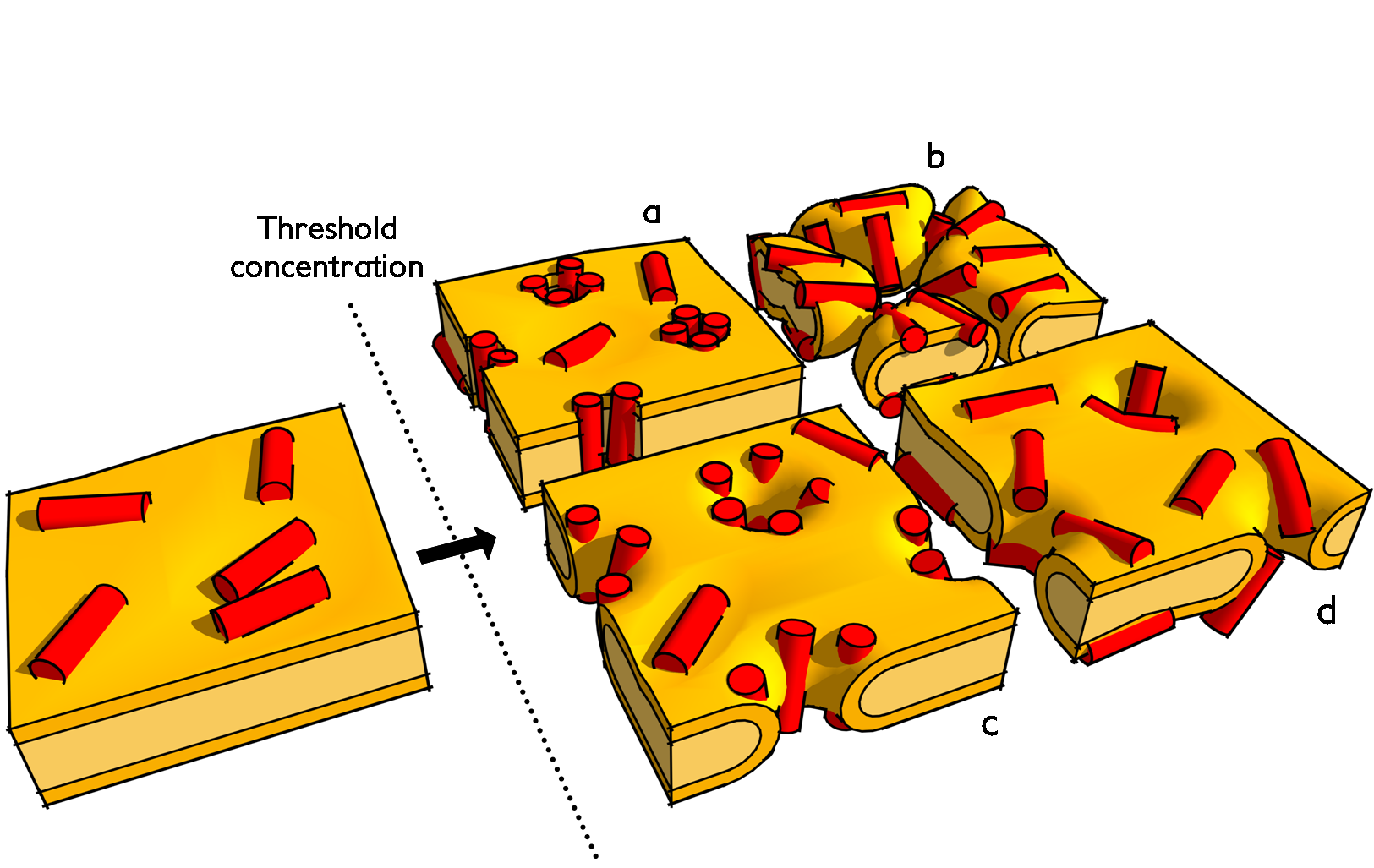
Proposed membrane-active mechanisms of antimicrobial peptides. Upon the crossing of a critical membrane-bound concentration the membrane is disrupted by either: a) barrel-stave pores; b) detergent-like carpeting; c) toroidal pores; or d) disordered toroidal pores.
Membrane lipid organization
Phospholipid membranes aren't always a homogeneous mixture of their components. Depending on the temperature and composition lipid domains of different order can form.
In complex lipid mixtures, such as the cell membrane, clear domain formation hasn't been observed. Rather, zones of enrichment and depletion of certain lipid types can be seen to develop.
The Martini model is particularly suited to studying these phenomena.

Top view of lipid tails phase separating in a mixed bilayer.
Mixtures of saturated-tail lipids (DPPC, in green) with unsaturated-tail lipids (DLiPC, in red) and cholesterol (in blue) separate into ordered and disordered domains at physiological temperatures.
Parameterization
The performance and accuracy of the Martini model undergo continuous improvement. Recent developments include the use of spatially constrained virtual interaction sites to increase simulation stability.
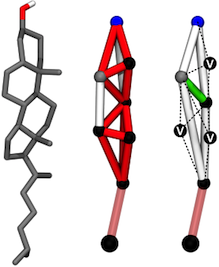
Atomistic and coarse-grained models of cholesterol.
Left: All-atom representation of cholesterol.
Center: Original Martini cholesterol model. The tridimensional structure was constrained through the use of multiple stiff bonds (in red) and bond constraints (in white).
Right: The improved Martini cholesterol model. The unstable network of bonds and constraints was replaced by two frames of constraints from where three virtual site beads (labeled 'V') are built.
Multiscaling
For the situations where more detail is needed than what the Martini model can afford multiscaling approaches can be used.
The AdResS scheme is a method to combine regions of different resolution in the same simulation box. This allows the speed-up of the coarse-grain region while retaining the detail of an atomistic region.
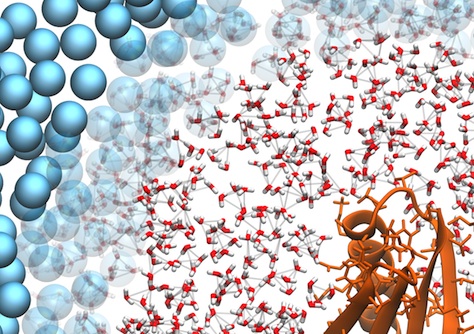
Detail of a multiscale simulation of a protein in water. The combination of coarse-grain and atomistic regions was done with the AdResS scheme.



