Extensin Peroxidases
Extensin Peroxidases: Structure and Function
The structural cell wall protein, extensin, is thought to play essential developmental roles in primary wall biosynthesis, cell adhesion, the definition of cell morphology and the regulation of extension growth. These 50-90 kDa glycoproteins can be readily isolated from some callus cultures as monomers, but become largely insoluble under normal conditions in planta, a process thought to involve the oxidative inter- and intra-cross-linking of extensin Tyrosine residues to form a 3D network. This structural protein network electrostatically associates and interweaves with other wall polymers, and it is in this network state that extensin exerts its major effects on wall (and tissue) properties. |
|
Identifying Extensin Peroxidases. Despite the fundamental role played by extensin in dicot plants, relatively little is known concerning the enzymatic basis of extensin cross-linking. The term 'Extensin Peroxidase' (EP) was coined by the group of Derek Lamport in the late 1980s to describe an anionic class III peroxidase from tomato callus culture with unusually high extensin cross-linking activity (Everdeen et al., 1988). Other lines of evidence, including the observation of extensin cross-linking in muro, and its inhibition by classical peroxidases inhibitors, suggest that a sub-group of class III peroxidases (EPs) mediate extensin cross-linking in planta.
Since then, a few 'EPs' have been found in other species. However, identifying EPs has proven problematic, even though EPs must be at least as wide-spread in higher plants (especially dicots) as extensin itself.
Part of the problem is that EP activity is conventionally assayed by monitoring extensin oligomerisation with gel-filtration chromatography, and therefore cannot provide precise parameters (Vmax, Km) of first order kinetics. The problem is further compounded by the use of differing reaction conditions by different researchers, which makes the comparison of 'EP activity' described by different laboratories impractical.
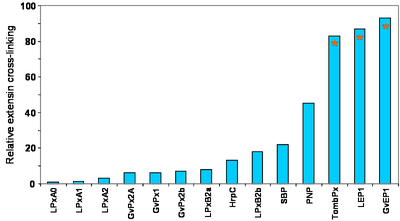
The PCW group has developed an assay for EP activity in muro to more closely simulate conditions in vivo. The classical peroxidase models; HrpC, SBP, PNP, as well as 12 peroxidases isloated from lupin, grapevine and tomato have been assayed, and clearly higher EP activities could be associated with GvEP1, LEP1 and TombEP from grapevine, lupin and tomato, respectively. PNP was also relatively efficient.
Fig. 2. In muro assay for EP activity suggests a cut-off to distinguish EPs from other class III peroxidases. The assay utilises 60 µg GvP1 extensin/ mg (Dwt) of saline-extracted grapevine callus cell walls. The assay was performed in 20 mM NaAcetate, pH 4.5, and initiated with 1 mM H2O2. Reaction time was 10 min and arrested by extensive washing in NaAcetate buffer.
|
|
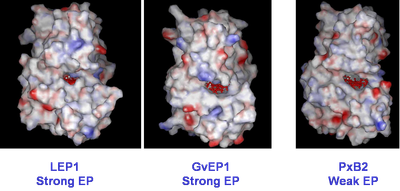
Extensin Peroxidase Structure. The demonstrable high capacity of a few peroxidases for extensin cross-linking could be promoted by their structural or chemical differences to other class III peroxidases. The identification of structural determinants for EP activity was therefore initiated by cloning EPs from lupin (Price et. al, 2003) and grapevine (Pereira et al. 2011; Jackson & Vatulescu, unpublished). Homology-based modelling was subsequently utilised to compare these enzymes with crystal structures of other class III peroxidases in the data-base. A grapevine peroxidase with weak EP activity, PxB2b (Fig. 2) was also included.
Fig. 3. Fig. 3. Homology-based modelling of two strong EPs; LEP1 and GvEP1. A grapevine peroxidase with weak EP activity was included for comparative purposes. The comparison indicated that an unobstructed equatorial cleft bordering the substrate access channel may be required for EP activity. However, an unobstructed access of macromolecules to the heme edge is clearly not the only pre-requisite for high EP activity, as seen by the presence of this feature in the non-EP, PxB2. |
|
To continue these studies, we have expressed the strong EP, GvEP1, in E.coli and are in the process of renaturing the enzyme activity. We intend to utilise RAMAN, NMR and crystallographic studies, to probe the interaction of this EP with small, extensin-like peptides.
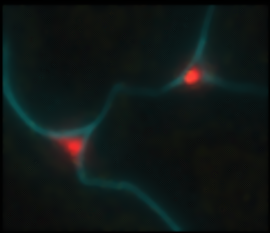
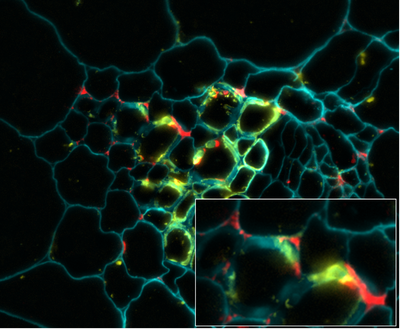
Immunolocalisiation of Grapevine Extensin Peroxidase 1. Although it might be reasonable to suppose that those class III peroxidases (EPs) with high extensin cross-linking activity are utilsed for such in planta, this pre-supposition has never been examined directly. We have therefore studied the localisation of both extensin and the EP, GvEP1, in developing grapevine tissues, using highly specific antibodies. The study also included the use of polyclonal anti-HrpC Abs, which can detect other peroxidase epitopes, and so allow the comparison of GvEP1 localisation with that of the general peroxidase population.
Fig. 4. Co-localisation of GvEP1 with extensin in developing vascular bundles of grapevine epicotyls. Both proteins are expressed in xylem wall faces and within cell corners of immediately surrounding xylem parenchyma (see inset). Whereas it might be expected that extensin epitopes are also localised in tissues free from GvEP1, the consistent co-localisation of GvEP1 with extensin epitopes indicates its functional association with extensin network formation in vivo. GvEP1 shows a considerably more restricted distribution than that of HrpC epitopes (data not shown). |
|