Cytochrome Oxidase
Proton transfer in Haem-Copper Oxidases
Collaboration between the Laboratories of Cláudio M. Soares and António M. Baptista.
Financed project: PTDC/QUI-BIQ/113446/2009 entitled "Proton transfer and proton pumping in haem-copper oxidases. Methodological developments and their application to unravel the molecular mechanism "
Haem-copper oxidases are at the core of the aerobic metabolism, receiving electrons from other redox proteins and catalyzing the reduction of oxygen to water. Concomitant with this, and together with the protons for oxygen reduction, other protons are pumped across the membranes where they are inserted, generating a proton gradient. This proton gradient can be used by ATP synthases to produce ATP.
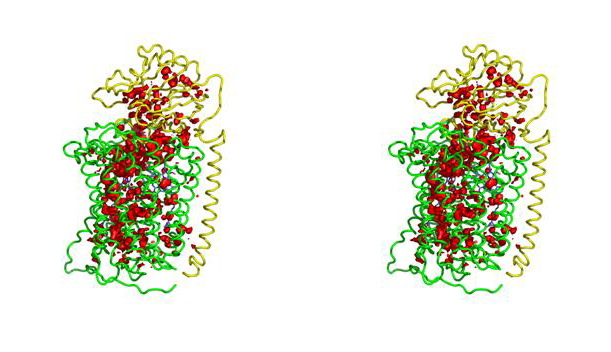
The structure of Paracoccus denitrificans aa3 haem-copper oxidase can be appreciated in the following figure.
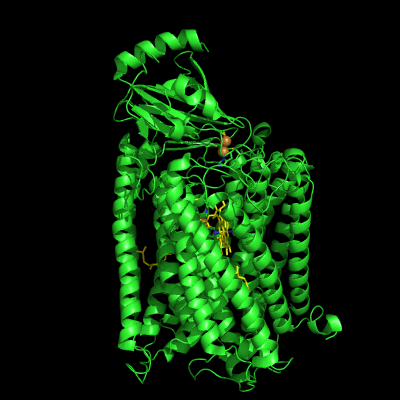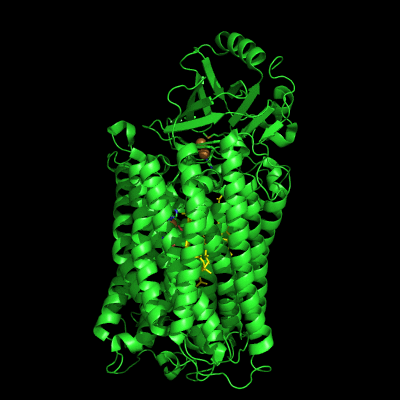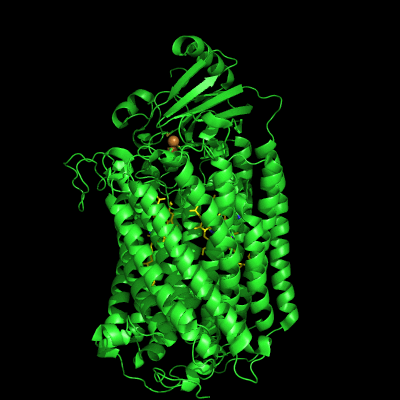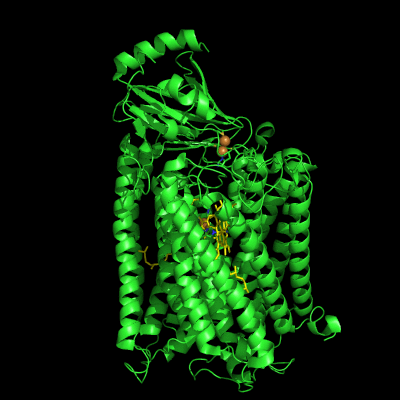
These systems pump protons from one side of the membrane to the other, giving rise to the proton gradient. The exact molecular details behind this process are still unknown, and researchers have suggested the existence of proton channels, constituted by protonatable and polar residues as well as water molecules, which would transfer the protons across the protein matrix. These has been, and will be, the object of our research.
Given that water molecules are important in defining proton channels we have investigated the water placement in model haem-copper oxidases, using new methods within the context of comparative models
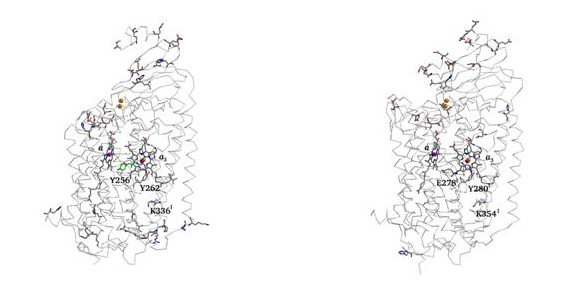
Finding protonatable groups at physiological pH is important into the identifications of possible proton channels. The following figure contains a representation of proton active groups found using this procedure, in two different haem-copper oxidases.
We are hiring. If you are interested in this project, please send us an email.
Recent relevant publications:
Victor, BL, Baptista, AM, Soares, CM (2004) "Theoretical identification of proton channels in the Quinol Oxidase aa3 from Acidianus ambivalens", Biophys.J., 87, 4316-25.
Soares, CM, Baptista, AM, Pereira, MM, Teixeira, M. (2004) "Investigation of protonatable residues in Rhodothermus marinus caa3 haem-copper oxygen reductase. Comparison with Paracoccus denitrificans aa3 haem-copper oxygen reductase", JBIC, 9, 124-134
Kirichenko, AV, Pfitzner, U, Ludwig, B, Soares, CM, Vygodina, TV, Konstantinov, AA (2005) “Cytochrome c oxidase as a calcium binding protein. Studies on the role of a conserved aspartate in helice XI-XII cytoplasmic loop in cation binding.”, Biochemistry, 44, 12391-12401.



