Haemagglutinin
Molecular modeling of hemagglutinin from Influenza Viruses
Collaboration between the Laboratories of Cláudio M. Soares and Miguel Castanho.
Financed project: PTDC/QUI-BIQ/114774/2009 “Membrane fusion mechanism of Influenza Hemagglutinin: a simulation and biophysical approach.”
Influenza viruses (IV) are responsible for worldwide outbreaks of flu. The most devastating viral infection in the 20th century was not caused by HIV, but by the Spanish influenza, which was derived from an IV found in birds. Furthermore, viruses from the 1957 and 1968 pandemics were hybrids between human and avian viruses, and given that humans did not have immunity to avian influenza, these pandemics had devastating consequences. More recently, the new strain of H1N1 IV, which is a combination of genes from swine, avian and human influenza, generated the 2009 flu pandemic outbreak, which began in Mexico, and rapidly spread all over the world, causing numerous deaths. Therefore, it is critical to pursue studies on conserved mechanisms of the virus life-cycle, i.e. mechanisms largely independent on the specific strain. Viral fusion mechanism is probably the most conserved step in influenza life cycle. Understanding IV fusion is creating an opportunity to do it into a therapeutic target, opening a gateway to new therapeutic approaches, still valid when future unavoidable flu outbreaks strike again, saving lives and improving the quality of life of patients world-wide.
IV are orthomyxovirus, and are classified into A, B and C types, based on the antigenic differences of internal proteins. Different strains of IV are named based on host of origin, geographic location, strain number, year of isolation, and specially on antigenic classification of hemagglutinin (HA) and neuraminidase (NA), two glycoproteins found on their lipid membrane, which are involved in pathogenicity and virulence.
![]()
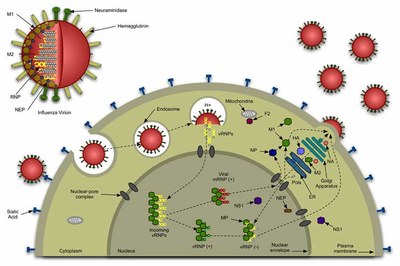
Influenza viruses life cycle
IV initiate infection by binding HA (a 231 kDa trimeric protein) to cell surface receptors containing sialic acid, entering the cell by receptor-mediated endocytosis. The low pH of the endosomes triggers a dramatic conformational change in HA (spring loaded model), exposing the fusion peptide (FP), which is responsible for the perturbation of the endosome membrane. In the end, these perturbation events and the major conformational alterations of HA pull the virus and endosome membranes together, allowing their fusion. This creates a pore that allows the delivery of the viral content into the cell cytoplasm, which is then used in the replication of the virus. After replication, new viruses are exocyted, ready to infect neighbour cells (see figure above).
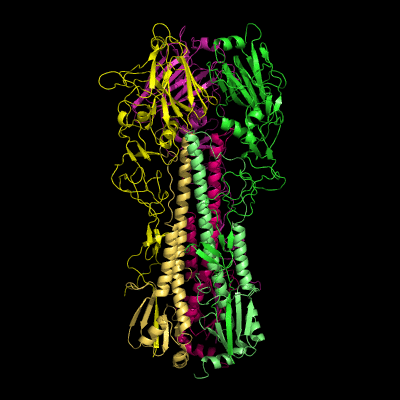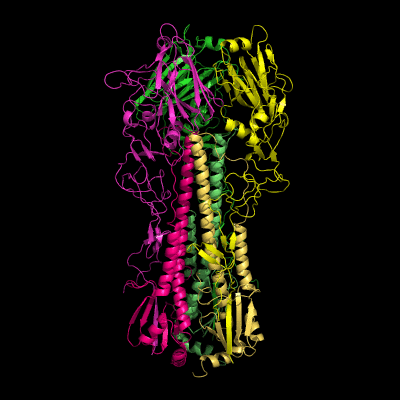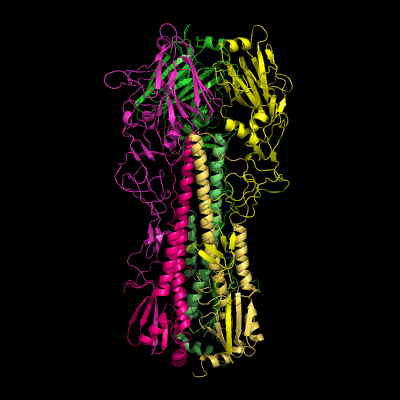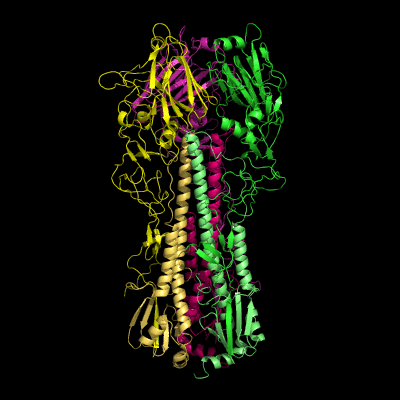
Hemagglutinin crystallographic structure
Each HA monomer is synthesized as a single chain of 550 resides, whose proteolytic cleavage produces two disulfide-bridged chains, HA1 and HA2. HA1 is hydrophilic and contains sialic acid receptor regions. Low pH induces a conformational change on this chain, which is accompanied by other structural changes in the HA2 chain, that in the end will allow the fusion peptide (FP) to be exposed, and thereby promoting membrane fusion. This peptide consists on the first 20-25 residues of the N-terminal of HA2, and is highly conserved in all types of IV strains, which makes fusion a unique target for therapies against potentially all strains, leading to a real advancement in medical care in the long term.
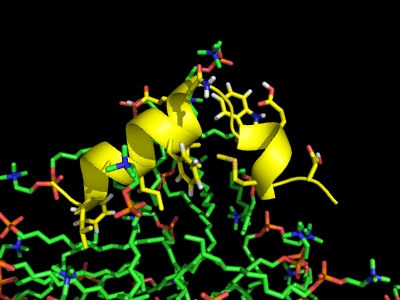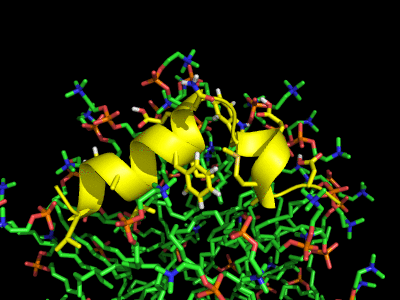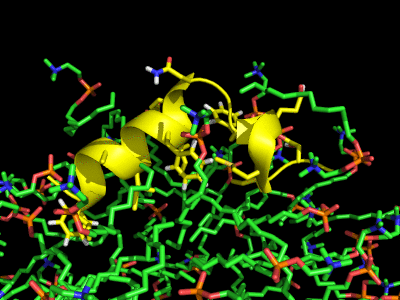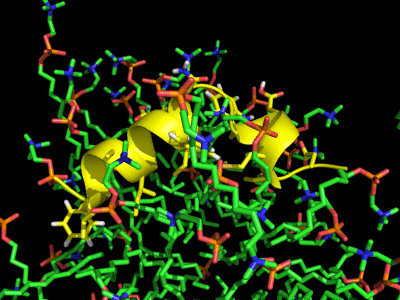
Simulation of the Fusion peptide
In this research project we aim to understand the structural determinants responsible for the major conformational changes that HA undergoes under low pH medium conditions, and that in the end will promote the fusion of the viral and endosomal membranes. This recently financed project consists in a collaboration between our laboratory and the Department of Biochemistry in the Institute of Molecular Medicine (IMM) (Prof. Miguel Castanho). By combining calculations with experiments, we will be able to provide a more detailed molecular picture and, at the same time, an experimental validation.
Recent relevant publications:
- Victor, BL, Baptista, AM, Soares, CM (2012) “Structural determinants for the membrane insertion of the transmembrane peptide of hemagglutinin from Influenza virus”, J.Chem. Infor. Model., accepted.



