Understanding protein structure and enzyme kinetics in non-aqueous solvents
Understanding protein structure and enzyme kinetics in non-aqueous solvents
Although water is the natural environment of enzymes, their use in nonaqueous solvents is of great technological and fundamental interest. In nonaqueous media, enzymes can display several novel and valuable properties, such as the capacity to catalyze reactions that are not feasible in water, increased stability, an altered substrate specificity as well as enantiomeric selectivity and molecular “memory”. In order to be able to take full advantage of this technological potential a solid knowledge of the enzymatic behavior in nonaqueous media is essential. Our aim is to contribute to a deeper understanding of the molecular determinants of nonaqueous enzymology, using molecular simulation techniques. Some of the questions we have addressed are:
• What is the structure and dynamics of proteins in an organic apolar solvent like?
• How do the solvent and its water content influence enantioselectivity?
• How is water distributed around the protein in polar and apolar solvents?
• What is the structure and dynamics of proteins in ionic liquids like?
• What are the structural determinants of ligand imprinting?
Molecular basis for cutinase enantioselectivity
In close collaboration with the experimental groups of Prof. Susana Barreiros (FCT/UNL) and Prof. Joaquim Sampaio Cabral (IST/UTL), we have contributed to the rationalization of the enantioselectivity of cutinase in nonaqueous solvents.
Cutinase: (R,S)-1-phenylethanol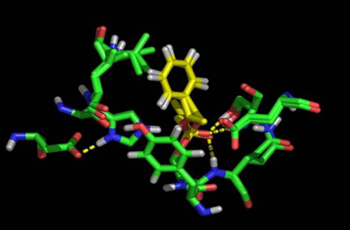 |
Cutinase: (R,S)-2-phenyl-1-propanol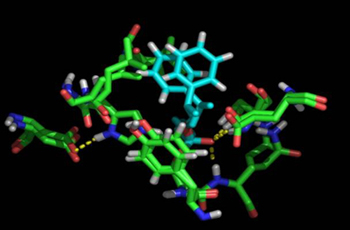 |
Molecular basis of rational discrimination between R,S enantiomers of 1-phenylethanol and 2-phenyl-1-propanol bound at Cutinase active site. R enantiomers are prefered by the enzyme. High enantioselectivity is experimentaly observed and computationaly modelled for the R-1-phenylethanol. The R-2-phenyl-1-propanol is also prefered but in less extent. Collaboration with the groups of Susana Barreiros and Sampaio Cabral.
Hydration mechanisms of enzymes in polar and nonpolar media
We have analyzed the hydration mechanisms of the serine protease cutinase in five organic solvents with different polarities (Micaelo and Soares (2007), FEBS J., 274, 2424-2436). Our results indicate that the amount, structure and dynamics of water at the enzyme surface are clearly dependent on the nature of the solvent. Nevertheless, as can be observed in the figure bellow, water seems is localized at equivalent regions of the enzyme surface independently of the organic solvent employed.
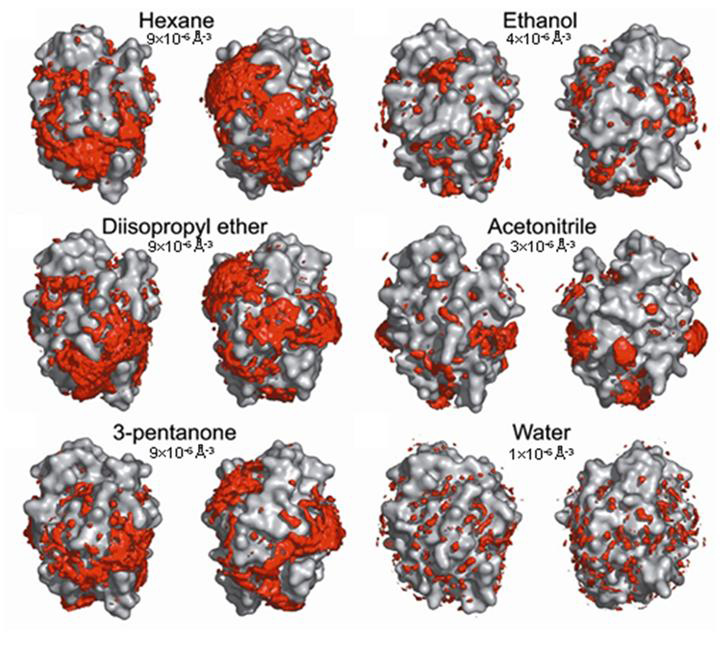
Spatial distribution of water around cutinase in polar and apolar solvents. For each organic solvent, two sides of the enzyme are shown in order to give a complete view of the surface. The contours enclose regions with a probability density above the value indicated in the figure (Micaelo N.M. and Soares C.M. (2007), FEBS J., 274, 2424-2436)
Protein structure in ionic liquids
The first MD simulation of an enzyme in ionic-liquids was performed in our laboratory (Micaelo and Soares (2008), J. Phys. Chem. B, 112, 2566-2572). In this work we have used the serine protease cutinase as a model enzyme in two ionic liquids with the same cation but different anions: [BMIM]-[NO3] and [BMIM][PF6]. One of the most interesting findings of this study was that the enzyme is less stabilized in [BMIM]-[NO3] relative to [BMIM][PF6], both at 298 K and 343 K, most likely due to the ability of the [NO3]-anion to form hydrogen bonds with the protein main chain, therefore disrupting intra-protein hydrogen bonds.
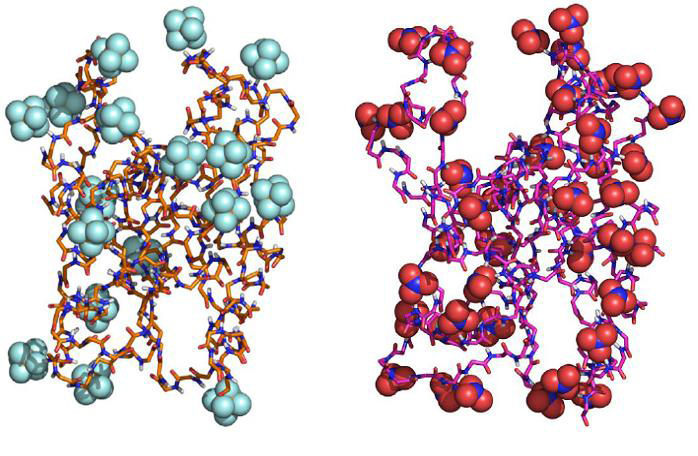
Final conformation of cutinase at 343 K with 2.5% water percentage in [BMIM][PF6] (left) and [BMIM]-[NO3] (right) (Micaelo and Soares (2008), J. Phys. Chem. B, 112, 2566-2572)
Structural determinants of ligand imprinting
Enzymes in organic solvents appear to “remember” the presence of a ligand after it has been removed. This phenomenon is known as ligand imprinting and is not observed in water. We have analyzed the molecular causes of ligand imprinting through a MD simulation approach, using the serine protease subtilisin as a model enzyme (Lousa et al (2011) Prot. Sci., 20, 379-386). In this study we started by docking a competitive inhibitor in the active site of subtilisin and then we performed 10 ns of MD simulation in water to adapt the active site of the enzyme to the ligand.
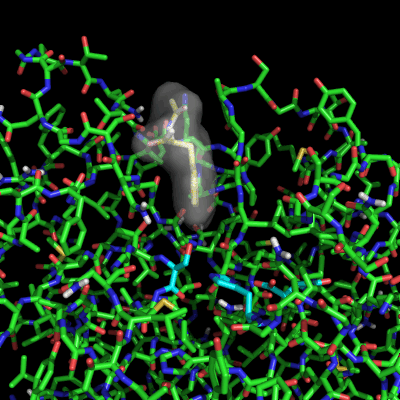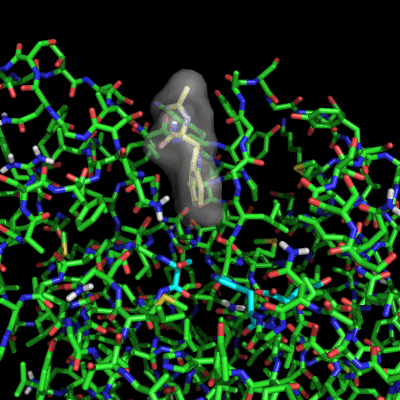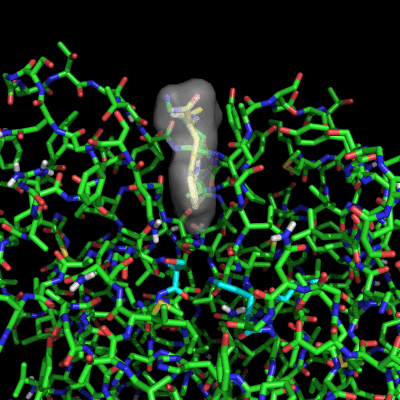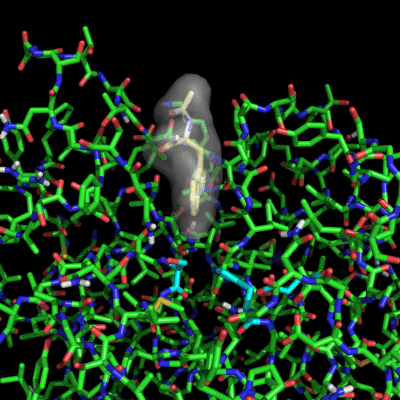
The next step was to remove the ligand and perform simulations both in water and in hexane. The movies bellow show that in hexane the pocket remains open, whereas in water it rapidly closes and adopts an intermediate state.
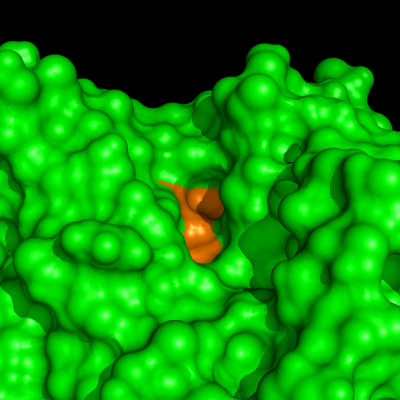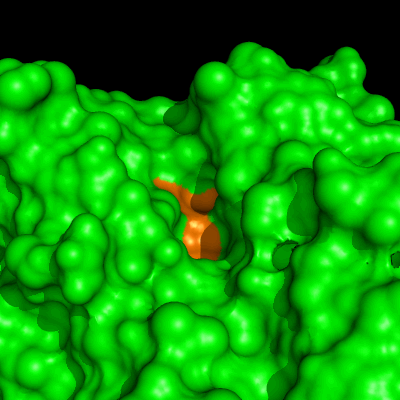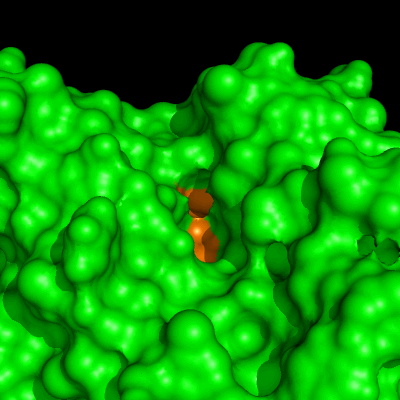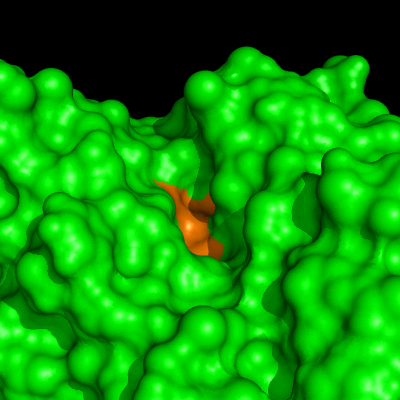
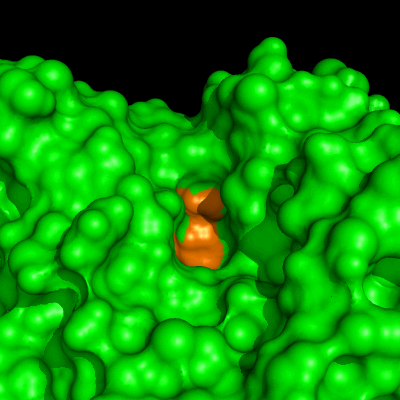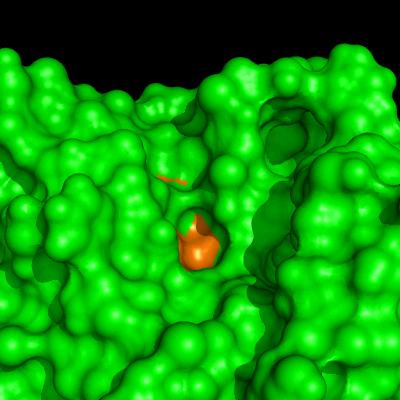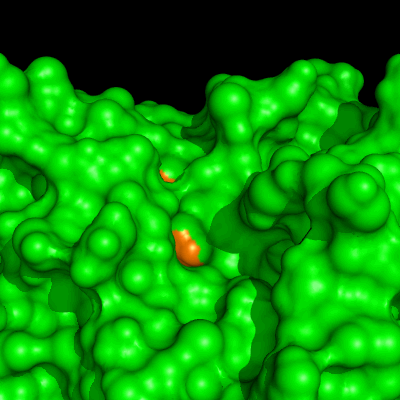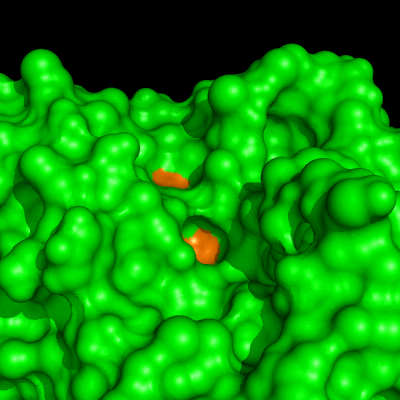
Relevant publications:
- Fontes, N., Almeida, M.C., Peres, C, Garcia, S., Grave, J., Aires-Barros, M.R., Soares, C.M., Cabral, J.M.S., Maycock, C.D., Barreiros, S. (1998) "Cutinase activity and enantioselectivity in supercritical fluids", Ind. Eng. Chem. Res., 37, 3189-3194
- Soares, C.M., Teixeira, V.H., Baptista, A.M. (2003) "Protein structure and dynamics in nonaqueous solvents: insights from molecular dynamics simulation studies", Biophys. J., 84, 1628-1641
- Vidinha, P., Harper, N., Micaelo, N.M., Lourenço, N.M., da Silva, M.D., Cabral, J.M., Afonso, C.A., Soares, C.M., Barreiros, S. (2004) "Effects of immobilization support, water activity, and enzyme ionization state on cutinase activity and enantioselectivity in organic media", Biotechnol Bioeng., 85, 442-449
- Micaelo, N.M., Teixeira, V.H., Baptista, A.M., Soares, C.M. (2005) “Water dependent properties of cutinase in nonaqueous solvents: A computational study of enantioselectivity”, Biophys. J., 89, 999-1008
- Micaelo, N.M., Baptista, A.M., Soares, C.M. (2006) “Parametrization of 1-butyl-3-methylimidazolium hexafluorophosphate/nitrate ionic liquid for the GROMOS force field”, J. Phys. Chem. B, 110, 14444-14451
- Micaelo, N.M., Soares, C.M. (2007) “Modeling hydration mechanisms of enzymes in nonpolar and polar organic solvents”, FEBS J., 274, 2424-2436
- Micaelo, N.M., Soares, C.M. (2008) “Protein structure and dynamics in ionic liquids. Insights from molecular dynamics simulation studies”, J. Phys. Chem. B, 112, 2566-2572
- Lousa, D., Baptista, A.M., Soares, C.M. (2011) “Structural determinants of ligand imprinting: A molecular dynamics simulation study of subtilisin in aqueous and apolar solvents”, Prot. Sci., 20, 379-386
- Lousa, D, Baptista, AM, Soares, CM (2012) “Analyzing the molecular basis of enzyme stability in ethanol/water mixtures using molecular dynamics simulations”, J.Chem.Infor.Model., 52: 465-473
- Lousa, D, Cianci, M, Helliwell, JR, Halling, PJ, Baptista, AM, Soares, CM (2012) “Interaction of counterions with subtilisin in acetonitrile: Insights from molecular dynamics simulations”, J.Phys.Chem.B, 116, 5838–5848



