Amidase
We have determined the 3D structure of an aliphatic amidase in complex with a reaction intermediate. This is a project in collaboration with Prof. Amin Karmali, ISEL, Lisbon, Portugal.
Microbial amidases belong to the thiol nitrilases family and have potential biotechnological applications in chemical and pharmaceutical industries as well as in bioremediation.The amidase from Pseudomonas aeruginosa is a 6x38-kDa enzyme that catalyzes the hydrolysis of a small range of short aliphatic amides. Its high resolution crystallographic structure showed that each amidase monomer is formed by a globular four-layer alpha-beta-beta-alpha sandwich domain with an additional 81-residue long C-terminal segment. This wraps arm-in-arm with a homologous C-terminal chain of another monomer, producing a strongly packed dimer. In the crystal, the biological active homo-hexameric amidase is built grouping three such dimers around a crystallographic 3-fold axis. The structure elucidates the structural basis for the enzyme activity, with the nitrilases catalytic triad at the bottom of a 13-Å deep, funnel-shaped pocket, accessible from the solvent through a narrow neck with 3-Å diameter. An acyl transfer intermediate, resulting from the purification protocol, was found bound to the amidase nucleophilic agent, Cys(166). The obtained structure suggest that some pocket defining residues should undergo conformational shifts to allow substrates and products to access and leave the catalytic pocket, for turnover to occur.
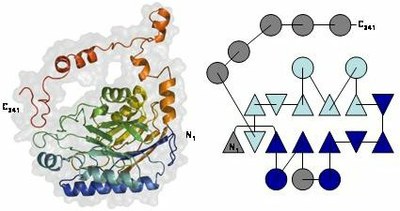
Amidase monomer shows four-layer abba-sandwich architecture with an extended C-terminal region including several helical segments. Its topology diagram (triangles representing b-strands and circles representing a-helices) evidences that the two mixed b-sheets (in dark and light blue) are topologically equivalent. The monomer results from their duplication (additional secondary-structure elements in gray). Superposition of the two monomer halves N- and Cstructural motifs, in light blue and red, respectively, also present in available structures of the nitrilase family.
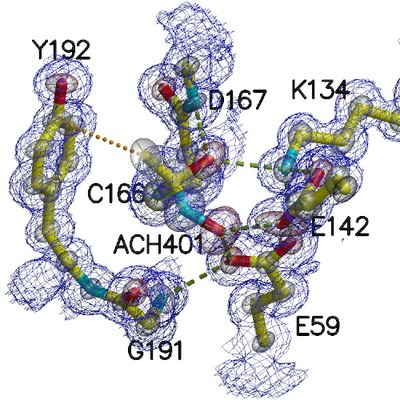
The high-resolution diffraction allowed a model refinement with anisotropic atomic displacement parameters (transparent ellipsoids) and provided new insights of amidase catalytic site: an acyl reaction intermediate (ACH401) was found establishing contacts not only with the catalytic triad Glu(59)-Lys(134)-Cys(166) but also with other active site conserved residues, which showed direct interactions with the reaction intermediate.
Andrade & Frazão et al. JBC 282,19598 (2007)



