Research Highlights
eHooke: a tool for automated image analysis of spherical bacteria based on cell cycle progression
Fluorescence microscopy is a critical tool for cell biology studies on bacterial cell division and morphogenesis. As the analysis of fluorescence microscopy images evolved beyond initial qualitative studies, numerous images analysis tools were developed to extract quantitative parameters on cell morphology and organization. To understand cellular processes required for bacterial growth and division, it is particularly important to perform such analysis in the context of cell cycle progression. However, manual assignment of cell cycle stages is laborious and prone to user bias. Although cell elongation can be used as a proxy for cell cycle progression in rod-shaped or ovoid bacteria, that is not the case for cocci, such as Staphylococcus aureus. We developed the image analysis framework eHooke, specifically for automated analysis of microscopy images of spherical bacterial cells. eHooke contains a trained artificial neural network to automatically classify the cell cycle phase of individual S. aureus cells. Users can then apply various functions to obtain biologically relevant information on morphological features of individual cells and cellular localization of proteins, in the context of the cell cycle.
eHooke: A tool for automated image analysis of spherical bacteria based on cell cycle progression. Saraiva BM, Krippahl L, Filipe SR, Henriques R, Pinho MG. Biol Imaging. 2021;1:e3. doi: 10.1017/S2633903X21000027.
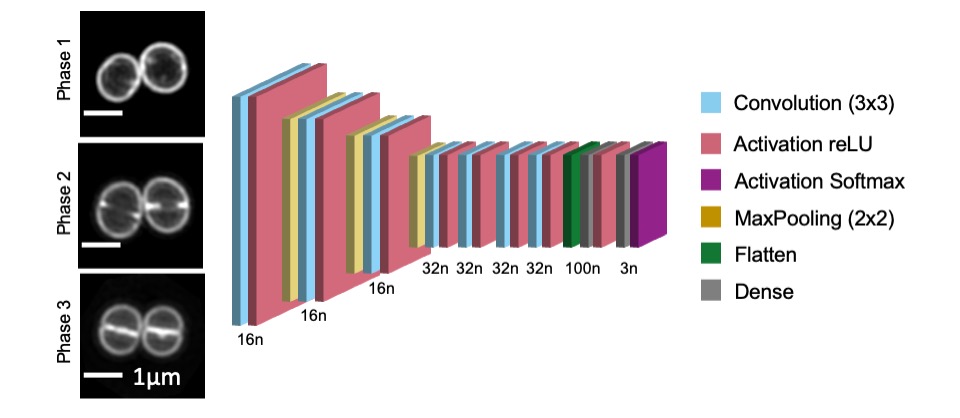
Image shows S. aureus JE2 cells stained with membrane dye Nile red and imaged by Structured Illumination Microscopy (SIM). From top to bottom are examples of cells in cell cycle Phase 1 (prior to initiation of division septum synthesis), Phase 2 (during which the septum is synthesized) and Phase 3 (during which cells have a complete septum that divides the mother cell in two, prior to splitting into two daughter cells). An artificial neural network with the structure represented above (where n represents the number of neurons in each layer) can be used to automatically classify the cell cycle phase of individual S. aureus cells.
Reassessment of the distinctive geometry of Staphylococcus aureus cell division.
Staphylococcus aureus was thought to divide in three alternating orthogonal planes over three consecutive division cycles. Although this mode of division was proposed over four decades ago, the molecular mechanism that ensures this geometry of division has remained elusive. In this work we showed that S. aureus cells do not regularly divide in three alternating perpendicular planes as previously thought. Imaging of the divisome showed that a plane of division is always perpendicular to the previous one, avoiding bisection of the nucleoid, which segregates along an axis parallel to the closing septum. However, one out of the multiple planes perpendicular to the septum which divide the cell in two identical halves can be used in daughter cells, irrespective of its orientation in relation to the penultimate division plane. Therefore, division in three orthogonal planes is not the rule in S. aureus as though for such a long time. For a movie on S. aureus cell division see here.
Reassessment of the distinctive geometry of Staphylococcus aureus cell division. Saraiva BM, Sorg M, Pereira AR, Ferreira MJ, Caulat LC, Reichmann NT, Pinho MG. Nat Commun. 2020;11:4097. doi: 10.1038/s41467-020-17940-9.
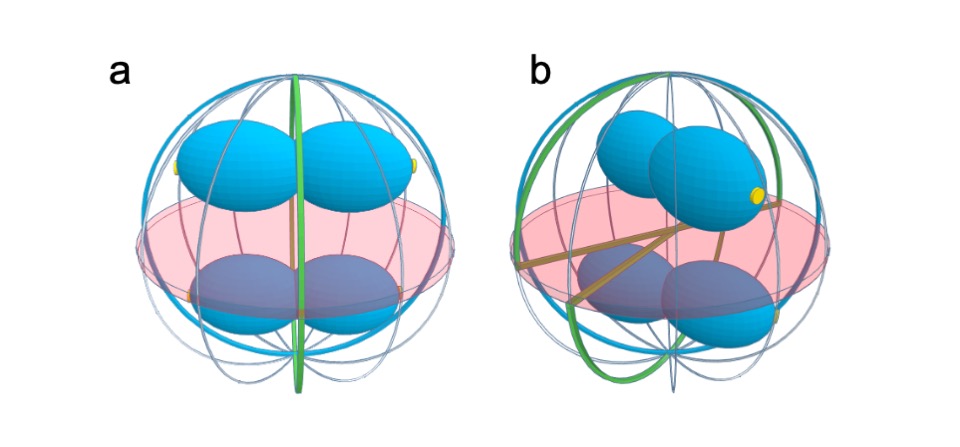
Scheme of dividing S. aureus cells showing the scar of the previous division plane (blue ring), the current division plane (red disk) and the localization of the future division plane, where the divisome assembles (green ring). Chromosomes are shown in blue with origins of replication shown as small yellow discs. a The axis of chromosome segregation, which is parallel to the closing septum, establishes the division plane in S. aureus, as the origin proximal region is bound by the nucleoid occlusion effector Noc, which prevents FtsZ assembly. The only division plane that does not lead to bisection of the nucleoid is the plane indicated in green. b Chromosome segregation towards any of the meridians (grey lines) is geometrically equivalent. Therefore, chromosomes in sister cells can segregate in different orientations, releasing different planes for division (green lines). If division occurred in three consecutive orthogonal planes, the same plane of division had to be selected in both daughter cells, given that these share the two previous division planes. If that is not the case, then S. aureus cells do not necessarily divide in three consecutive orthogonal planes over three division cycles.
SEDS-bPBP pairs direct Lateral and Septal Peptidoglycan Synthesis in Staphylococcus aureus
Bacteria are a very diverse group of organisms and that can have various shapes, including spherical (cocci) or cylindrical (rods). Bacterial cell shape is maintained by directing incorporation of peptidoglycan, the major component of the bacterial cell wall, to distinct regions of the cell, namely through the localisation of late stage peptidoglycan synthesis proteins. These include two key protein families, SEDS transglycosylases and bPBP transpeptidases, proposed to function in cognate pairs. Rod-shaped bacteria have two SEDS-bPBP pairs, involved in elongation and division. In this paper, we elucidate why coccoid bacteria, such as Staphylococcus aureus, thought to lack an elongation machinery, also possess two SEDS-bPBP pairs. We determined that S. aureus RodA-PBP3 and FtsW-PBP1 constitute cognate pairs of interacting proteins. Lack of RodA-PBP3 resulted in more spherical cells due to deficient sidewall peptidoglycan synthesis, whereas depletion of FtsW-PBP1 arrested normal septal peptidoglycan incorporation. We propose that the FtsW-PBP1 pair has a role in stabilising the division machinery at midcell. In the absence of these proteins, the divisome appears as multiple rings/arcs that drive lateral peptidoglycan incorporation, leading to cell elongation. We conclude that RodA-PBP3 and FtsW-PBP1 mediate sidewall and septal PGN incorporation, respectively, and that their activity must be balanced to maintain coccoid morphology.
Reichmann NT, Tavares AC, Saraiva BM, Jousselin A, Reed P, Pereira AR, Monteiro JM, VanNieuwenhze MS, Fernandes F, Pinho MG. SEDS-bPBP pairs direct lateral and septal peptidoglycan synthesis in Staphylococcus aureus. Nature Microbiology. 2019; 4:1368-1377.doi: 10.1038/s41564-019-0437-2.
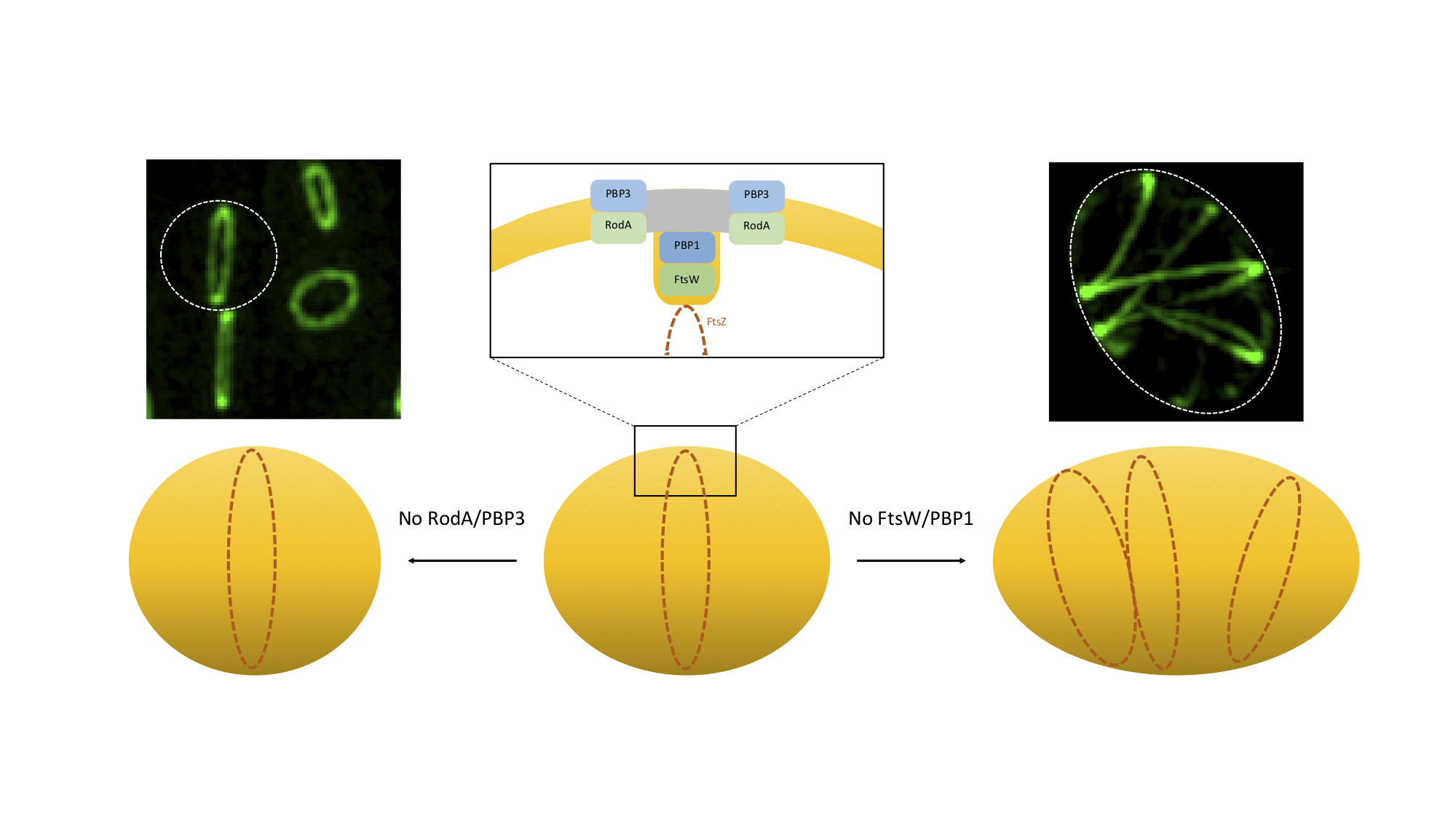
Model for the role of SEDS/bPBPs in Staphylococcus aureus morphogenesis.
S. aureus cells slight elongation depends on the activity of RodA/PBP3, which catalyse side-wall synthesis of peptidoglycan at the future division site (grey). In the absence of this cognate pair, staphylococcal cells become rounder. FtsW/PBP1 have a dual function during cell division. This pair is required for septal peptidoglycan synthesis, possibly to reorient the synthesis of peptidoglycan from the lateral wall to the perpendicular septum, promoting inward growth of the septum, as well as to stabilise the divisome. In the absence of FtsW/PBP1 the divisome is no longer localised exclusively at midcell, but forms multiple rings/arcs, capable of promoting PGN synthesis, leading to cell elongation. Microscopy images show a fluorescent derivative of divisome protein EzrA in the presence (left) or absence (right) of SEDS protein FtsW, visualized by Structured Illumination Microscopy (SIM).
Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis.
Peptidoglycan is the main component of the bacterial wall and protects cells from the mechanical stress that results from high intracellular turgor. When a bacterial cell divides, it has to synthesize peptidoglycan to form a septum, which will split the mother cell in two identical daughter cells in a process called cytokinesis. The molecular mechanism driving this process was unclear, with two competing models for the origin of the force drives cytokinesis: is it provided by the cytoskeleton, more specifically by the protein FtsZ, which is the bacterial homologue of tubulin? Or is it provided by the synthesis of peptidoglycan, which can be considered as the bacterial exoskeleton? In this paper we propose a model where S. aureus peptidoglycan synthesis machinery continuously incorporates peptidoglycan over the entire cell surface during the initial stage of the cell cycle, but is redirected to midcell in preparation for division. This causes massive peptidoglycan synthesis activity at the leading edge of the constricting septum which drives cytokinesis from that point onwards. Our data indicates that the two current models proposed in the literature for the origin of the force driving cytokinesis can be reconciled. We propose that cytokinesis occurs in two steps: an initial, slow one, driven by FtsZ and that may be responsible for the initial invagination of the cell membrane, followed by a second, faster step, for which peptidoglycan synthesis provides the driving force.
Monteiro JM, Pereira AR, Reichmann NT, Saraiva BM, Fernandes PB, Veiga H, Tavares AC, Santos M, Ferreira MT, Macário V, VanNieuwenhze MS, Filipe SR, Pinho MG. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. 2018. Nature. 554:528-532. doi: 10.1038/nature25506.
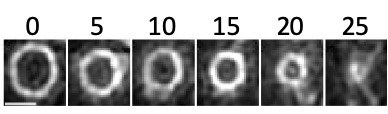
Cytokinesis in Staphylococcus aureus
Timelapse images, acquired every 5 minutes, showing constriction of the FtsZ ring during cytokinesis. The cell division protein FtsZ was fused to a fast folding version of the fluorescent protein GFP and visualized using Structured Illumination Microscopy (SIM). Time in minutes; scale bar 500nm.
Staphylococcus aureus requires at least one FtsK/SpoIIIE protein for correct chromosome segregation
Faithful coordination between bacterial cell division and chromosome segregation in rod-shaped bacteria, such as Escherichia coli and Bacillus subtilis, is dependent on the DNA translocase activity of proteins from the FtsK/SpoIIIE familiy, which move DNA away from the division site before cytokinesis is completed. However, the role of these proteins in chromosome partitioning had not been well studied in spherical bacteria. In this paper we showed that the two Staphylococcus aureus FtsK/SpoIIIE homologues, operate in independent pathways to ensure correct chromosome segregation during cell division. SpoIIIE forms foci at the center of the closing septum in at least 50% of the cells that are close to complete septum synthesis. FtsK is a multifunctional septal protein with a C-terminal DNA translocase domain that is not required for correct chromosome segregation in the presence of SpoIIIE. However, lack of both SpoIIIE and FtsK causes severe nucleoid segregation and morphological defects, showing that the two proteins have partially redundant roles in S. aureus.
Veiga, H. & Pinho, M. G. Staphylococcus aureus requires at least one FtsK/SpoIIIE protein for correct chromosome segregation. 2017. Mol Microbiol. 103, 387-565. doi: 10.1111/mmi.13572
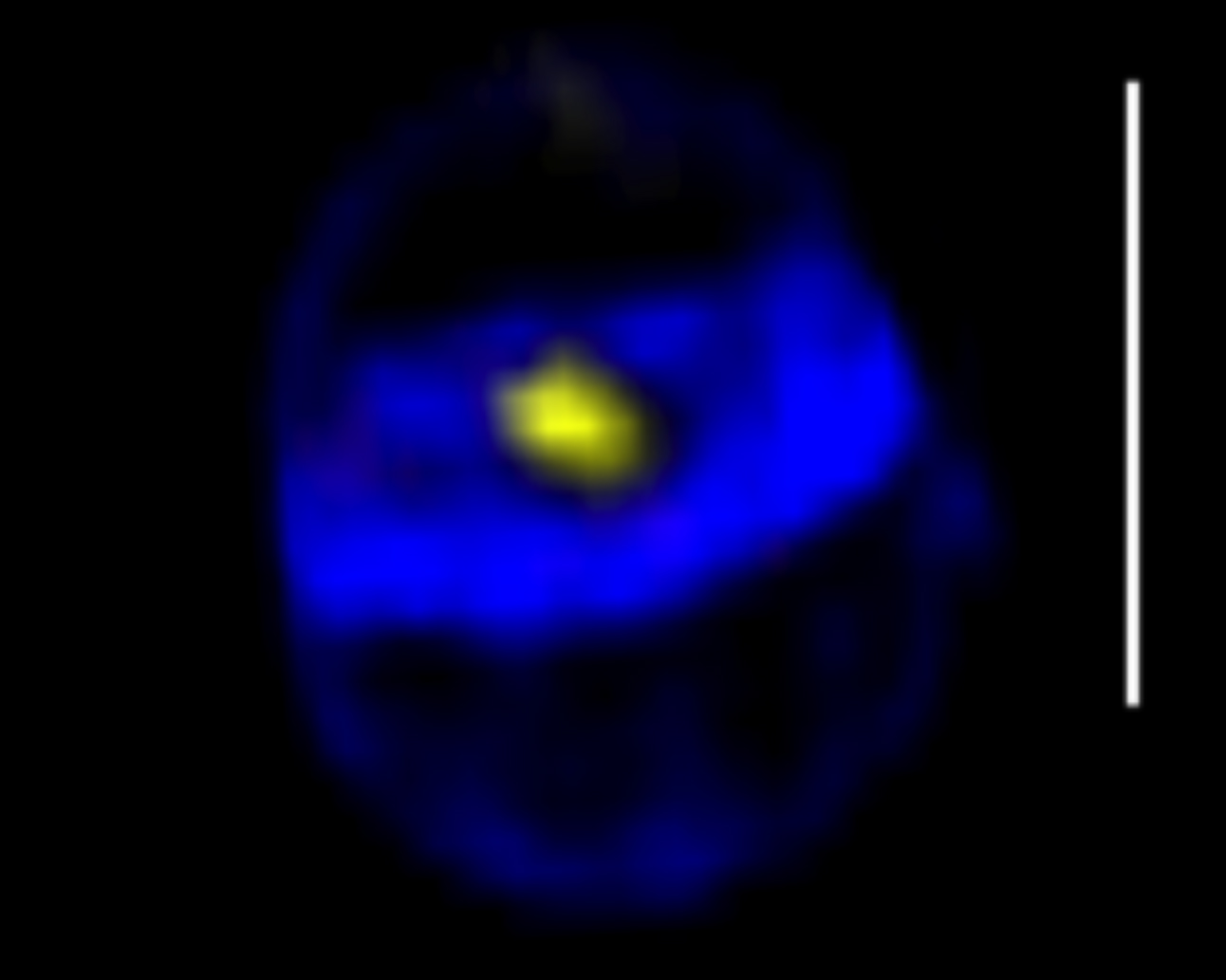
S. aureus SpoIIIE assembles in foci at the center of closing septa
Projection of a 3D-SIM image of a cell close to completion of septum synthesis, with a SpoIIIE-YFP foci in the center (yellow), showing that the SpoIIIE foci are formed before septum closure. The cell wall was labeled with the fluorescent aminoacid HADA. Scale bar 1 µm
FtsZ-dependent elongation of a coccoid bacterium
The mechanisms by which bacteria generate and maintain even the simplest cell shape remain an elusive but fundamental question in microbiology. In the absence of examples of coccus-to-rod transitions, the spherical shape has been suggested to be an evolutionary dead end in morphogenesis. In this paper we described the first observation of the generation of elongated cells from truly spherical cocci, occurring in a Staphylococcus aureus mutant containing a single point mutation in its genome, in the gene encoding the bacterial tubulin homologue FtsZ. We demonstrate that FtsZ-dependent cell elongation is possible, even in the absence of dedicated elongation machinery.
Pereira AR, Hsin J, Król E, Tavares AC, Flores, P, Hoiczyk E, Ng N, Dajkovic A, Brun, YV, VanNieuwenhze, MS, Roemer, T, Carballido-Lopez, R, Scheffers, D-J, Huang, KC and Pinho MG FtsZ-dependent elongation of a coccoid bacterium. 2016. MBio. 7:e00908-16. doi: 10.1128/mBio.00908-16.
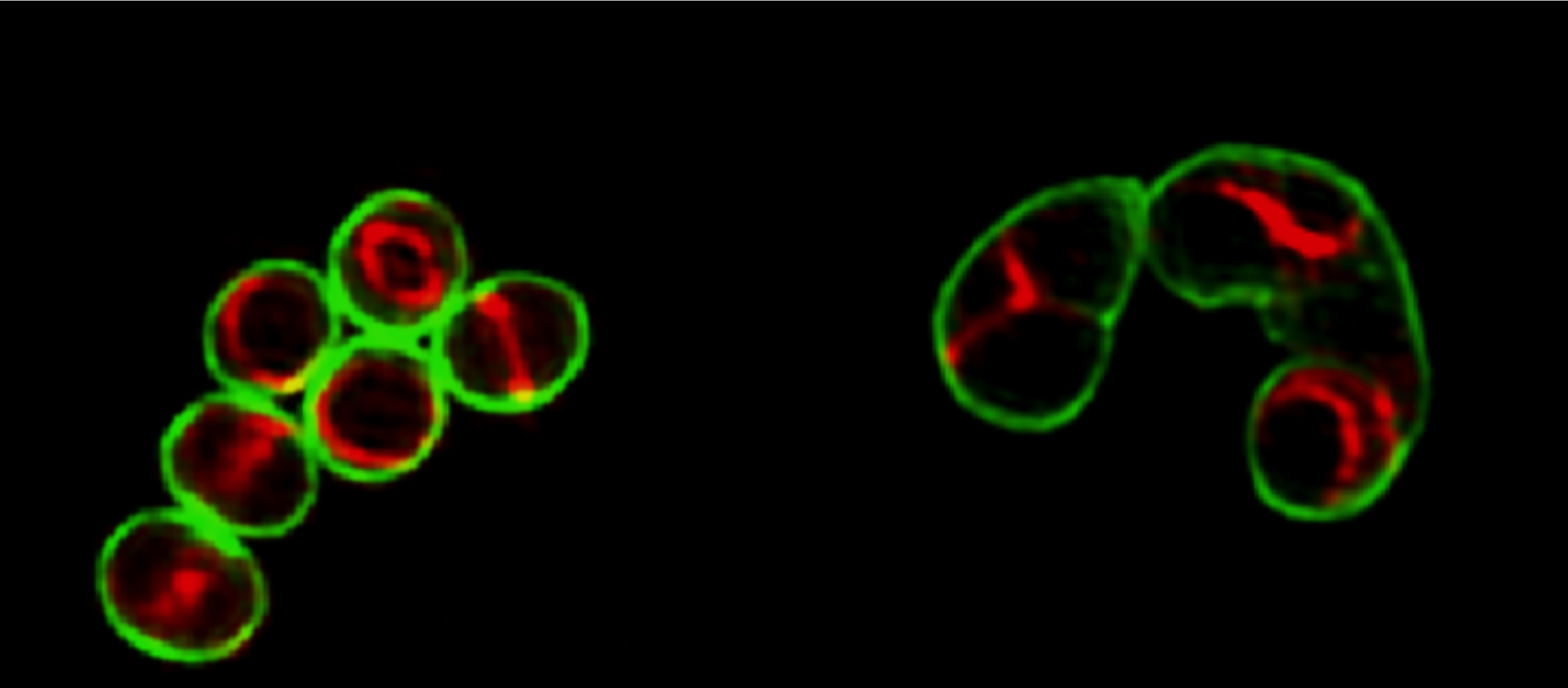
Elongating Staphylococcus aureus
Structured illumination microscopy (SIM) images of wild-type S. aureus (left) and an FtsZG193D mutant (right) expressing a fluorescent derivative of the FtsZ-associated protein EzrA-mCherry (red), labeled with the cell-wall dye Vancomycin-FL (green).
MreC and MreD proteins are not required for growth of Staphylococcus aureus.
The transmembrane proteins MreC and MreD are present in a wide variety of bacteria and are thought to be involved in cell shape determination. Together with the actin homologue MreB and other morphological elements, they play an essential role in the synthesis of the lateral cell wall in rod-shaped bacteria. In ovococci (shaped as a rugby ball), which lack MreB homologues, MreCD are also essential and have been implicated in peripheral cell wall synthesis. In this work we addressed the possible roles of MreC and MreD in the spherical pathogen Staphylococcus aureus. We showed that MreC and MreD are not essential for cell viability and do not seem to affect cell morphology, cell volume or cell cycle control. MreC and MreD localize preferentially to the division septa, but do not appear to influence peptidoglycan composition, nor the susceptibility to different antibiotics and to oxidative and osmotic stress agents. Surprisingly, our results suggest that, contrary to what happens in other bacteria, the function of MreCD in S. aureus is not critical for cell division and cell shape determination.
Tavares, A.C., Fernandes, P.B., Carballido-López, R, Pinho, M.G. MreC and MreD Proteins Are Not Required for Growth ofStaphylococcus aureus.2015.PLoS One.10:e0140523. doi: 10.1371/journal.pone.0140523.
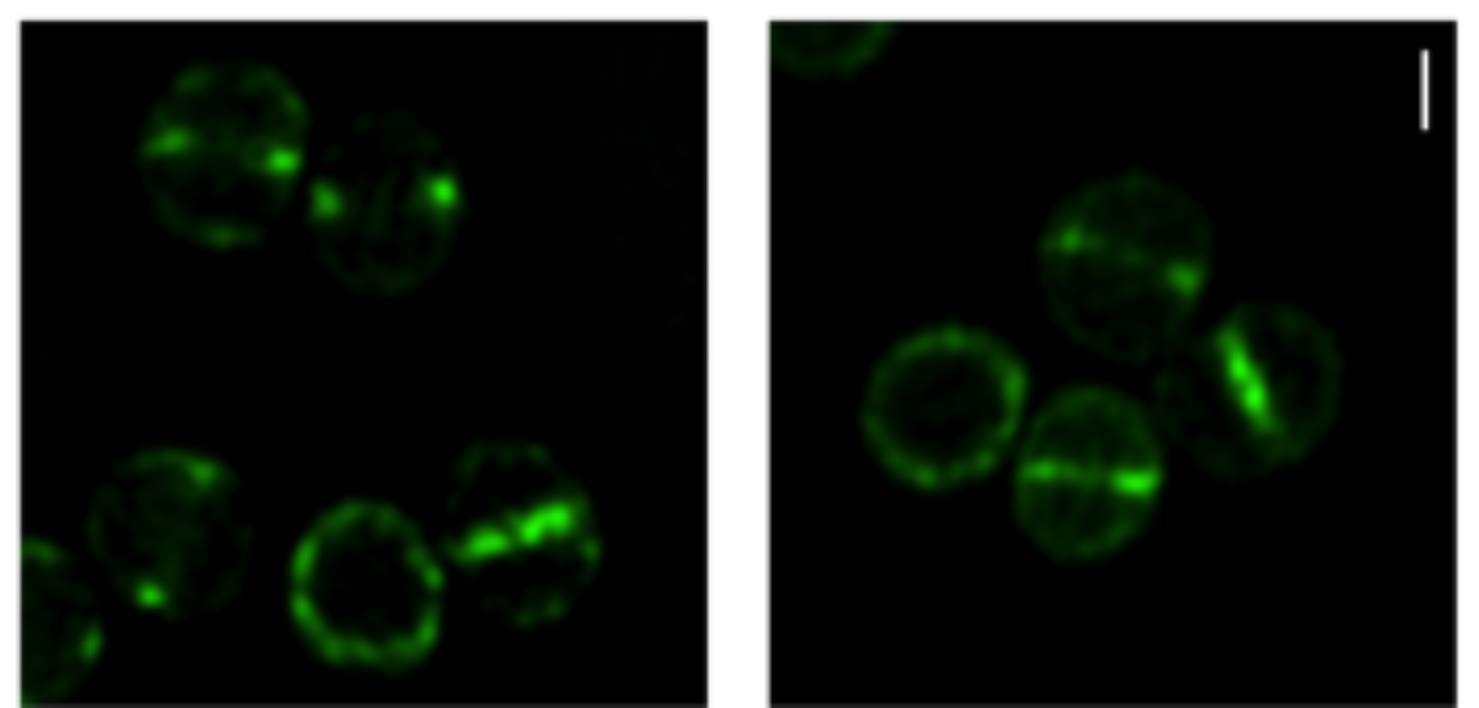
MreC and MreD are enriched at the division septum.
Localization of MreCD proteins
S. aureus COL cells expressing GFP fused to the N-terminal of MreC (left) or MreD (right) observed by super resolution Structured illumination microscopy (SIM). Scale bar, 0.5 μm.
Cell shape dynamics during the staphylococcal cell cycle.
Staphylococcus aureus is an aggressive pathogen and a model organism to study cell division in sequential orthogonal planes in spherical bacteria. However, the small size of staphylococcal cells has impaired analysis of changes in morphology during the cell cycle. In this work, we have used super-resolution microscopy to determine that S. aureus cells are not spherical throughout the cell cycle, but elongate during specific time windows, through peptidoglycan synthesis and remodeling. Both peptidoglycan hydrolysis and turgor pressure are required during division for reshaping the flat division septum into a curved surface. In this process, the septum generates less than one hemisphere of each daughter cell, a trait we showed is common to other cocci. Therefore, cell surface scars of previous divisions do not divide the cells in quadrants, generating asymmetry in the daughter cells. Our results introduce a need to reassess the models for division plane selection in cocci.
Monteiro JM, Fernandes PB, Vaz F, Pereira AR, Tavares AC, Ferreira MT, Pereira PM, Veiga H, Kuru E, VanNieuwenhze MS, Brun YV, Filipe SR, Pinho MG. Cell shape dynamics during the staphylococcal cell cycle. 2015. Nat. Commun. 6:8055. doi: 10.1038/ncomms9055
The cell cycle of Staphylococcus aureus
S. aureus COL cells were labeled with peripheral cell wall dye WGA-488 (green), washed, stained with membrane dye Nile Red and grown on the microscope slide.
Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance.
Many important cellular processes are performed by molecular machines, composed of multiple proteins that physically interact to execute biological functions. An example is the bacterial peptidoglycan (PG) synthesis machine, responsible for the synthesis of the main component of the cell wall and the target of many contemporary antibiotics. One approach for the identification of essential components of a cellular machine involves the determination of its minimal protein composition. Staphylococcus aureus is a Gram-positive pathogen, renowned for its resistance to many commonly used antibiotics and prevalence in hospitals. Its genome encodes a low number of proteins with PG synthesis activity (9 proteins), when compared to other model organisms, and is therefore a good model for the study of a minimal PG synthesis machine. We deleted seven of the nine genes encoding PG synthesis enzymes from the S. aureus genome without affecting normal growth or cell morphology, generating a strain capable of PG biosynthesis catalyzed only by two penicillin-binding proteins, PBP1 and the bi-functional PBP2. However, multiple PBPs are important in clinically relevant environments, as bacteria with a minimal PG synthesis machinery became highly susceptible to cell wall-targeting antibiotics, host lytic enzymes and displayed impaired virulence in a Drosophila infection model. The fact that S. aureus can grow and divide with only two active PG synthesizing enzymes shows that most of these enzymes are redundant in vitro and identifies the minimal PG synthesis machinery of S. aureus. However a complex molecular machine is important in environments other than in vitro growth, as the expendable PG synthesis enzymes play an important role in the pathogenicity and antibiotic resistance of S. aureus.
Reed P, Atilano ML, Alves R, Hoiczyk E, Sher X, Reichmann NT, Pereira PM, Roemer T, Filipe SR, Pereira-Leal JB, Ligoxygakis P, Pinho MG. Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance. 2015. PLoS Pathog. 11:e1004891. doi: 10.1371/journal.ppat.1004891.
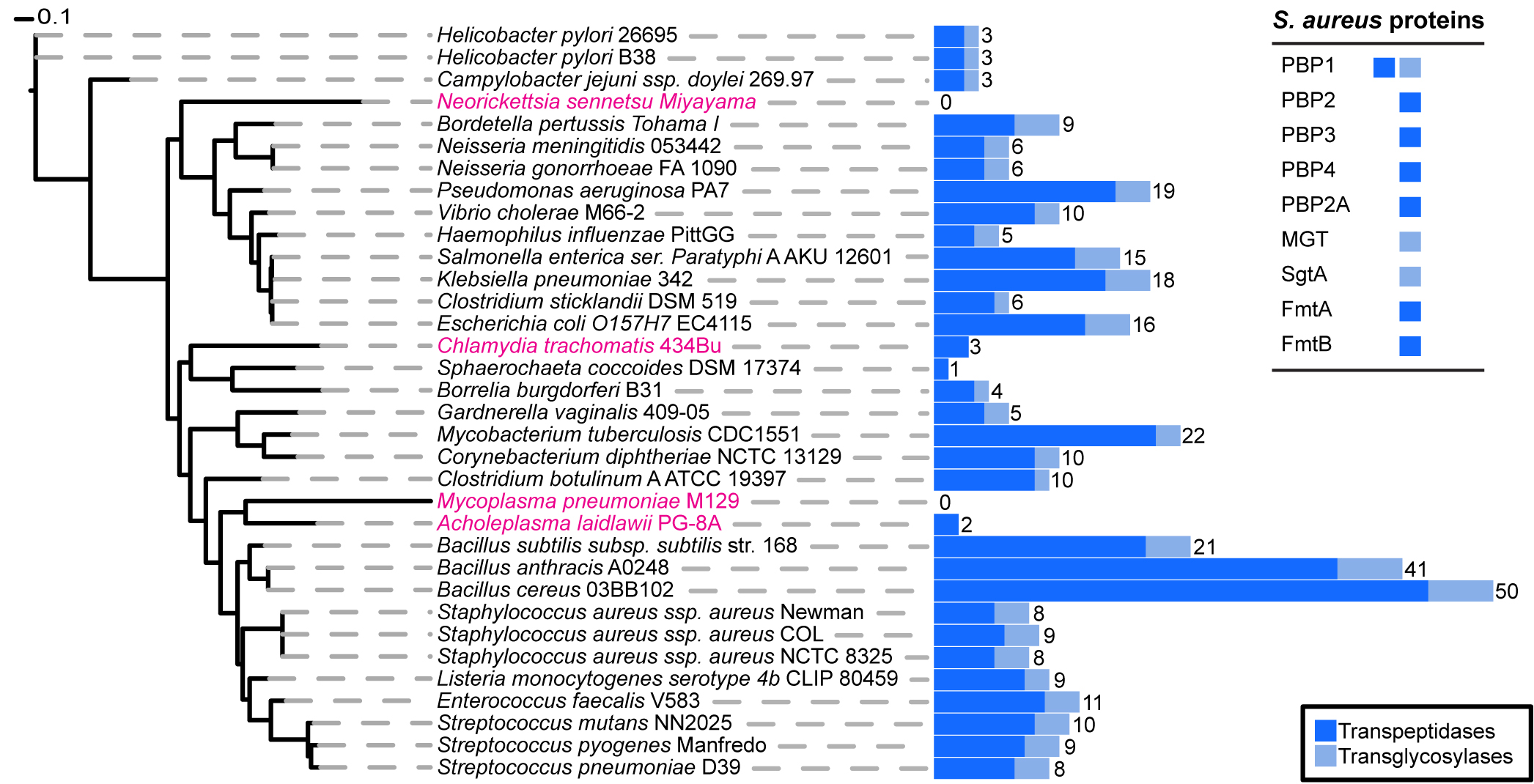
Phylogenetic distribution of peptidoglycan synthesis enzymes across selected bacterial species.
Blue bars represent the total number of proteins present in each species that have a transpeptidase (dark blue) or transglycosylase (light blue) domain. The total number of proteins that have at least one of the two domains is displayed numerically. Strains highlighted in pink are bacteria reported not to possess cell wall, although they may produce small but functional amounts of peptidoglycan. The table on the upper right corners shows the peptidoglycan synthesis proteins from S. aureus and their established or hypothetical activities. Seven of these nine proteins (all except PBP1 and PBP2) can be removed from S. aureus without impairing viability.
Characterization of a novel small molecule that potentiates β-lactam activity against gram-positive and gram-negative pathogens.
We contributed to a study led by Ambrose Cheung, Geisel School of Medicine at Dartmouth, USA, to characterize the mode of action of a new molecule with antibacterial activity, DNAC-1. This molecule was found in a loss-of-viability screen using small molecules against methicillin-resistant Staphylococcus aureus (MRSA) strain USA300, using a sub-inhibitory concentration of a β-lactam antibiotic. DNAC-1 potentiated the effect of oxacillin, i.e., the minimum inhibitory concentration (MIC) of oxacillin decreased from 64 to 0.25 μg/ml. Fluorescence microscopy indicated a disruption in the membrane structures within 15 min of exposure to DNAC-1 at 2× MIC. This permeabilization was accompanied by a rapid loss of membrane potential, as monitored by use of the DiOC2 dye. Macromolecular analysis showed the inhibition of cell wall synthesis by DNAC-1. Using a murine subcutaneous coinjection model, DNAC-1 alone or DNAC-1 with a sub-MIC of oxacillin resulted in a 6-log reduction in bacterial load and decreased abscess formation compared to the untreated control. DNAC-1 is a viable candidate in the development of combination therapy against many common bacterial pathogens.
D.R. Nair, J.M. Monteiro, G. Memmi, J. Thanassi, M. Pucci, J. Schwartzman, M.G. Pinho, A. Cheung. Characterization of a novel small molecule that potentiates β-lactam activity against Gram-positive and Gram-negative pathogens. 2015. Antimicrob Agents Chemother. 59:1876-1885. doi: 10.1128/AAC.04164-14.
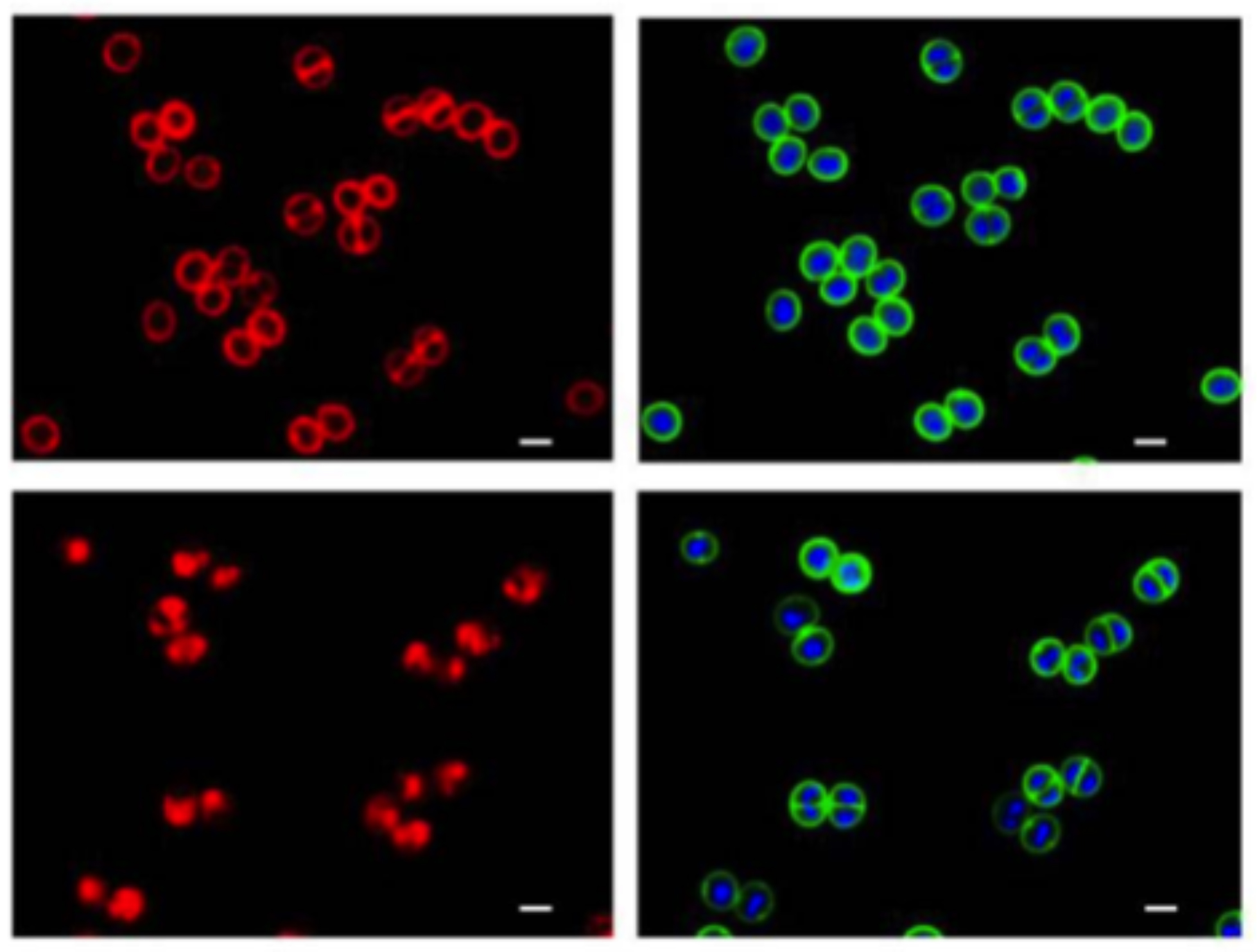
Disruption of cell membrane by DNAC-1 .
The cells were either untreated (top) or treated with 2× MIC of DNAC-1 for 15 min (bottom) and stained with FM 4-64 to visualize the membrane (left) and Bodipy-FL-vancomycin (green) to visualize the cell wall and Hoechst 33342 dye (blue) to see the DNA (right).
Reduction of the peptidoglycan crosslinking causes a decrease in stiffness of the Staphylococcus aureus cell envelope.
In collaboration with the group of Karin Jacobs, from Saarland University, Germany, we have used atomic-force microscopy (AFM) to probe the effect of peptidoglycan crosslinking reduction on the elasticity of the Staphylococcus aureus cell wall. Penicillin-binding protein 4 (PBP4) is a nonessential transpeptidase, required for the high levels of peptidoglycan crosslinking characteristic of S. aureus. Importantly, this protein is essential for β-lactam resistance in community-acquired, methicillin-resistant S. aureus (MRSA) strains but not in hospital-acquired MRSA strains. Using AFM we observed that the absence of PBP4, and the concomitant reduction of the peptidoglycan crosslinking, resulted in a reduction in stiffness of the S. aureus cell wall. Importantly, the reduction in cell wall stiffness in the absence of PBP4 was observed both in community-acquired and hospital-acquired MRSA strains, indicating that high levels of peptidoglycan crosslinking modulate the overall structure and mechanical properties of the S. aureus cell envelope in both types of clinically relevant strains.
P. Loskill, P.M. Pereira, P. Jung, M. Bischoff, M. Herrmann, M.G. Pinho* and K. Jacobs*. Reduction of the peptidoglycan crosslinking causes a decrease in stiffness of the Staphylococcus aureus cell envelope. 2014. Biophysical Journal. 107:1082-1089. * Both authors contributed equally to this work doi: http://dx.doi.org/10.1016/j.bpj.2014.07.029
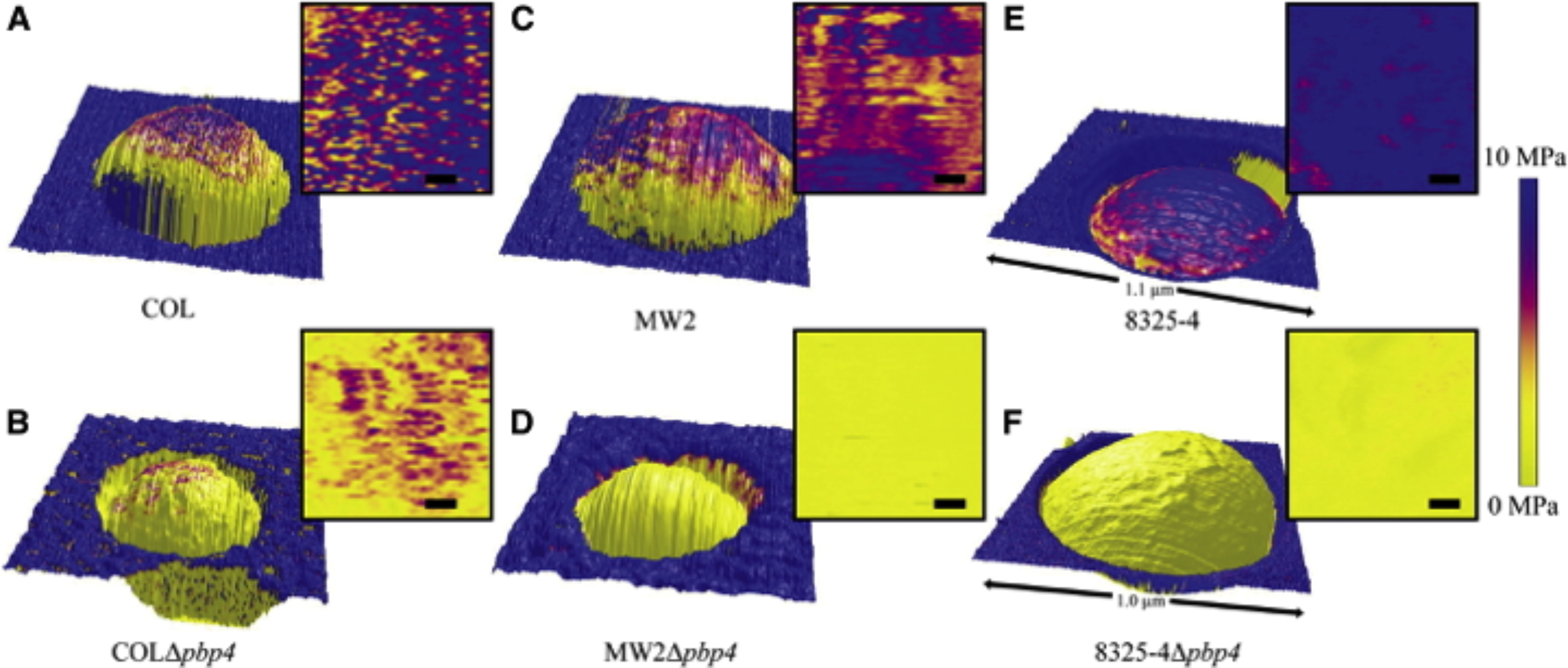 Height images overlaid with the distribution map of the elastic modulus values of (A) COL, (B) COLΔpbp4, (C) MW2, ( D) MW2Δpbp4, (E) 8325-4, and (F) 8325-4Δpbp4 single cells trapped in a membrane pore, and respective two-dimensional, top-down representations of the analyzed part of the elastic moduli (scale bars 50 nm). To avoid artifacts due to topography, only the topmost part (∼300 × 300 nm2) of each bacterium was analyzed. The stiffness of the cell wall of the Δ pbp4 mutants is significantly decreased in comparison with the corresponding wild-type strains.
Height images overlaid with the distribution map of the elastic modulus values of (A) COL, (B) COLΔpbp4, (C) MW2, ( D) MW2Δpbp4, (E) 8325-4, and (F) 8325-4Δpbp4 single cells trapped in a membrane pore, and respective two-dimensional, top-down representations of the analyzed part of the elastic moduli (scale bars 50 nm). To avoid artifacts due to topography, only the topmost part (∼300 × 300 nm2) of each bacterium was analyzed. The stiffness of the cell wall of the Δ pbp4 mutants is significantly decreased in comparison with the corresponding wild-type strains.
Review - How to get (a)round: mechanisms controlling growth and division of coccoid bacteria.
Bacteria come in a range of shapes, including round, rod-shaped, curved and spiral cells. This morphological diversity implies that different mechanisms exist to guide proper cell growth, division and chromosome segregation. Although the majority of studies on cell division have focused on rod-shaped cells, the development of new genetic and cell biology tools has provided mechanistic insight into the cell cycles of bacteria with different shapes, allowing us to appreciate the underlying molecular basis for their morphological diversity. In this Review, we discuss recent progress that has advanced our knowledge of the complex mechanisms for chromosome segregation and cell division in bacteria which have, deceptively, the simplest possible shape: the cocci.
Pinho MG, Kjos M, Veening JW. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. ( 2013) Nat Rev Microbiol. 11:601-614. doi: 10.1038/nrmicro3088
Inhibition of wall teichoic acid synthesis blocks the cooperative action of PBPs and sensitizes MRSA to β -Lactams
We collaborated in work led by Eric Brown, McMaster University, Canada to study the synergy between inhibition of wall teichoic acids (WTA) synthesis and peptidoglycan synthesis. WTA are anionic glycopolymers covalently attached to peptidoglycan, on the surface of gram-positive bacteria
Rising drug resistance is limiting treatment options for infections by methicillin-resistant Staphylococcus aureus (MRSA). One alternative for new treatments is the combination of two drugs, one which re-sensitizes resistant bacteria to β-lactams and a β-lactam antibiotic.
This work provides new evidence that WTA biogenesis is a remarkable antibacterial target with the capacity to destabilize the cooperative action of penicillin-binding proteins that is required for β-lactam resistance in MRSA. Deletion of gene tarO, encoding the first step of WTA synthesis, resulted in the restoration of sensitivity of MRSA to a unique profile of β-lactam antibiotics with a known selectivity for penicillin binding protein 2 (PBP2). Of these, cefuroxime was used as a probe to screen for previously approved drugs with a cryptic capacity to potentiate its activity against MRSA. Ticlopidine, the antiplatelet drug Ticlid, strongly potentiated cefuroxime, and this synergy was abolished in strains lacking tarO. The molecular target of ticlopidine was found to be the N-acetylglucosamine-1-phosphate transferase encoded by tarO gene. This provides evidence that WTA biogenesis represents an Achilles heel supporting the cooperative function of PBP2 and PBP4 in creating highly cross-linked muropeptides in the peptidoglycan of S. aureus. This approach represents a new paradigm to tackle MRSA infection.
M.A. Farha, A.Leung, E.W.Sewell, M.A.D'Elia, S.E.Allison, L.Ejim, P.M.Pereira, M.G.Pinho, G.D.Wright, E.D.Brown 2013. Inhibition of WTA Synthesis Blocks the Cooperative Action of PBPs and Sensitizes MRSA to β-Lactams. ACS Chem Biol. ACS Chem Biol. 8:226-233. doi: 10.1021/cb300413m.
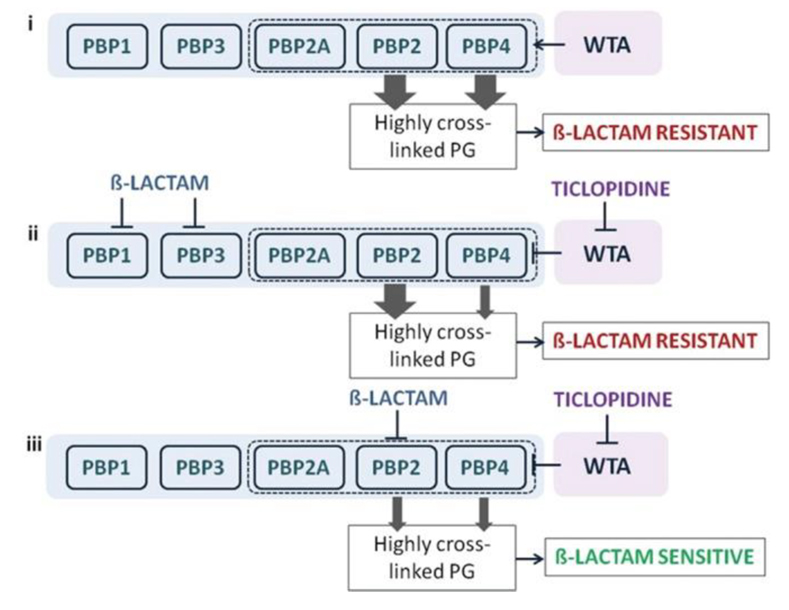
A synthetic lethal interaction when targeting TarO and native PBP2 .
Indirect inhibition of PBP4, via inhibition of TarO, and inhibition of PBP2 function account for the synergistic interaction among ticlopidine and cefuroxime. (i) When not challenged with β-lactams, WTA synthesis will guide the proper localization of PBP4 and, together with PBP2, will provide highly cross-linked muropeptide species (thick arrow) that contribute to high-level β-lactam resistance. (ii) Treatment with a β-lactam with low affinity for PBP2 and ticlopidine leads to an additive interaction as sufficient highly cross-linked peptidoglycan are present to maintain β-lactam resistance (one thick arrow), even when PBP4 function is affected by the lack of WTA (one thin arrow). (iii) Due to their cooperative function, PBP4 (when challenged with ticlopidine) and PBP2 (when challenged with β-lactam with high affinity for PBP2), will be impaired in their capacities to produce a highly cross-linked cell wall (two thin arrows), contributing to enhanced β-lactam susceptibility and thus a synergistic interaction.
Restoring methicillin-resistant Staphylococcus aureus susceptibility to beta-lactam antibiotics
We collaborated in work led by Terry Roemer, Merck Research Laboratories, USA to study the synergy between an FtsZ inhibitor and β-lactams.
Despite the need for new antibiotics to treat drug-resistant bacteria, current clinical combinations are largely restricted to β-lactam antibiotics paired with β-lactamase inhibitors. We have adapted a Staphylococcus aureus antisense knockdown strategy to genetically identify the cell division Z ring components-FtsA, FtsZ, and FtsW-as β-lactam susceptibility determinants of methicillin-resistant S. aureus (MRSA). We demonstrate that the FtsZ-specific inhibitor PC190723 acts synergistically with β-lactam antibiotics in vitro and in vivo and that this combination is efficacious in a murine model of MRSA infection. Fluorescence microscopy localization studies reveal that synergy between these agents is likely to be elicited by the concomitant delocalization of their cognate drug targets (FtsZ and PBP2) in MRSA treated with PC190723. A 2.0 Å crystal structure of S. aureus FtsZ in complex with PC190723 identifies the compound binding site, which corresponds to the predominant location of mutations conferring resistance to PC190723. Combining PC190723 with the β-lactam antibiotic imipenem markedly reduced the spontaneous frequency of PC190723-resistant mutants. Multiple MRSA PC190723 resistant FtsZ mutants also displayed attenuated virulence and restored susceptibility to β-lactam antibiotics in vitro and in a mouse model of imipenem efficacy. Collectively, these data support a target-based approach to rationally develop synergistic combination agents that mitigate drug resistance and effectively treat MRSA infections.
C.M. Tan, A.G. Therien, J. Lu, S.H. Lee, A. Caron,C.J. Gill, C. Lebeau-Jacob, L. Benton-Perdomo, J.M. Monteiro, P.M. Pereira, N. Elsen, J. Wu, K. Deschamps, M. Petcu, S. Wong, E. Daigneault, S. Kramer, L. Liang, E. Maxwell, D. Claveau, J. Vaillencourt, K. Skorey, J. Tam, H. Wang, T.C. Meredith, S. Sillaots, L. Wang-Jarantow, J.C. Reid, G. Parthasarathy, S. Sharma, A. Baryshnikova, K.J. Lumb, M.G. Pinho, S.M. Soisson, T.Roemer. Restoring Methicillin-resistant Staphylococcus aureus Susceptibility to beta-lactam Antibiotics. 2012. Science Translational Medicine. 4:126ra35. doi: 10.1126/scitranslmed.3003592.
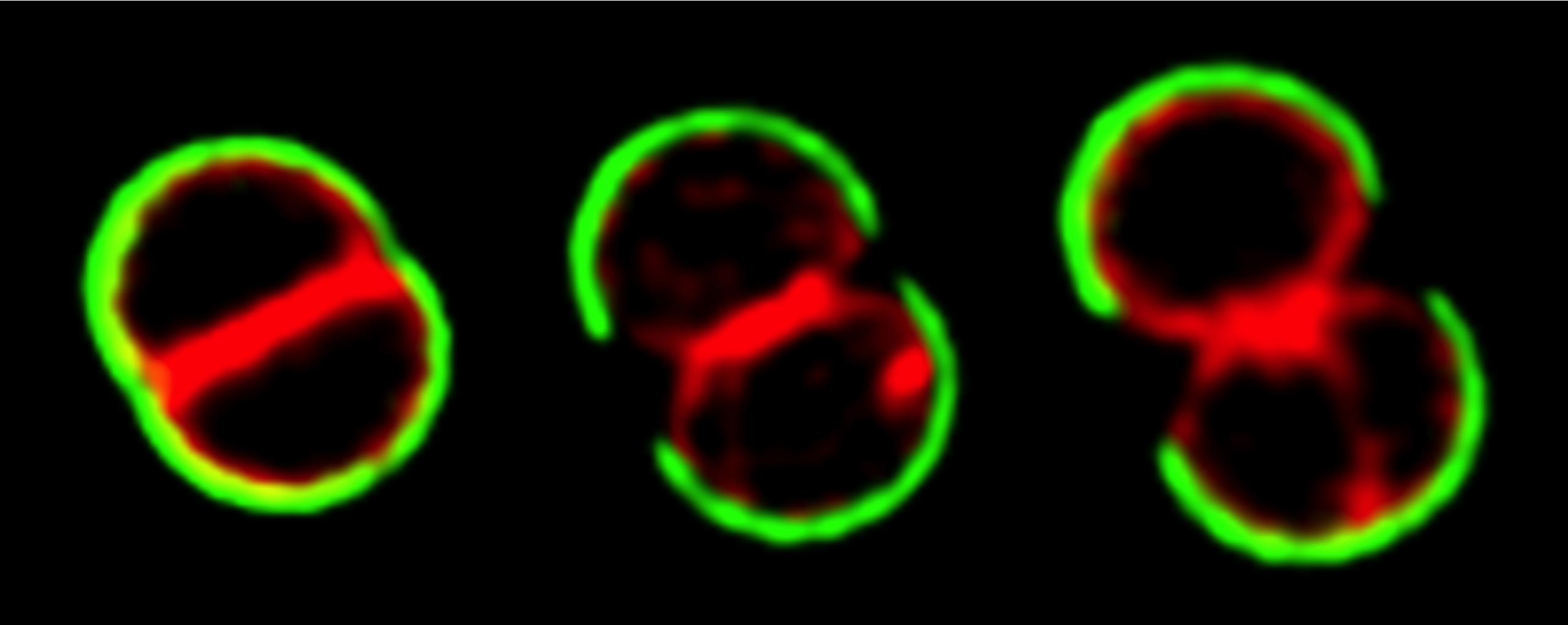
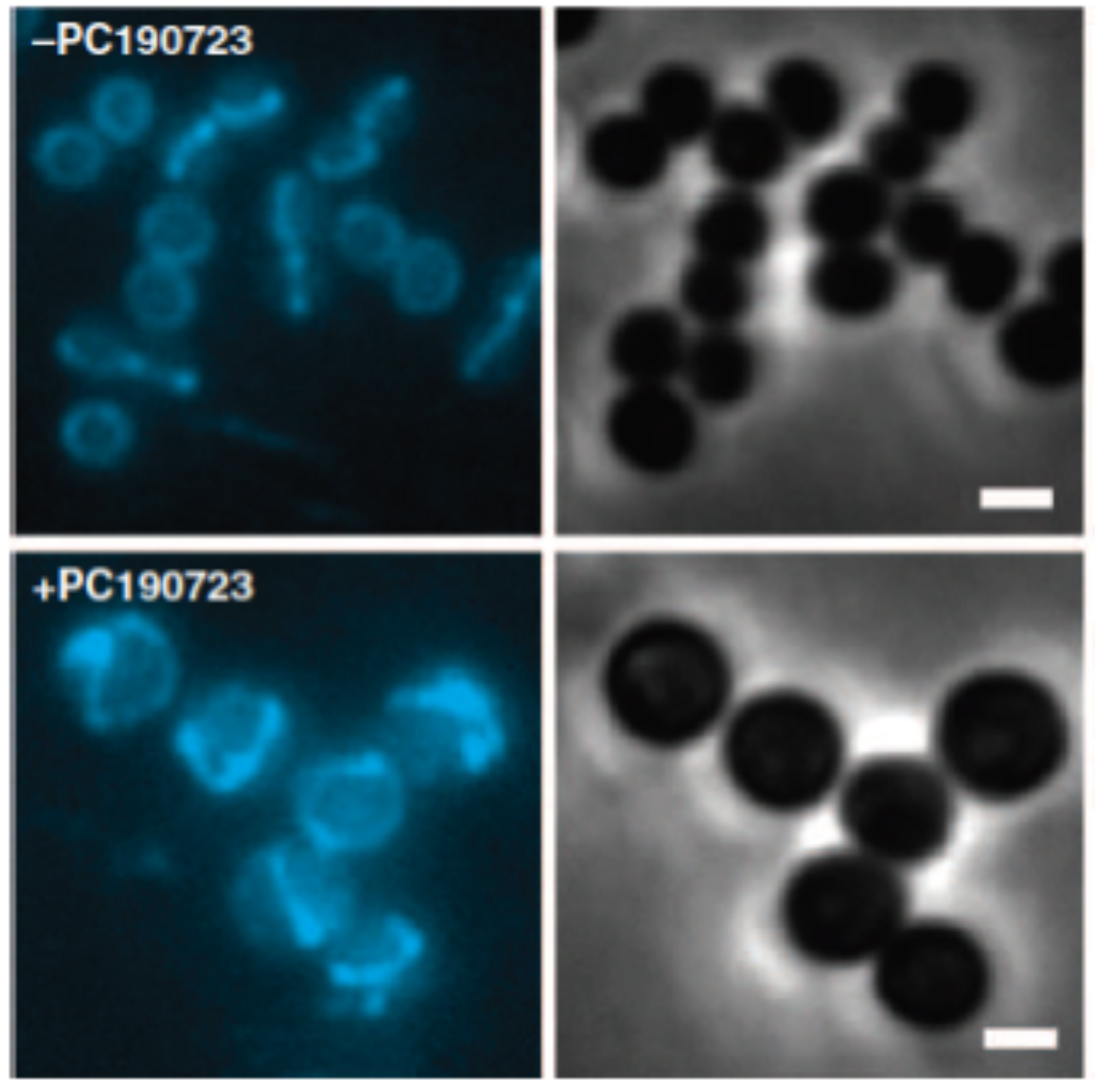 Septal localization of fluorescent derivatives of FtsZ is lost in PC190723-treated MRSA cells.
Septal localization of fluorescent derivatives of FtsZ is lost in PC190723-treated MRSA cells.
MRSA COL cells expressing FtsZ-CFP were grown in the absence (top panels) or presence of PC190723 (10 mg/ml) (bottom panels) for 30 min and analyzed by fluorescence microscopy. FtsZ-CFP fluorescence localizes to the division site in control cells versus multiple rings and arcs of FtsZ-CFP fluorescence in drug-treated cells. Right panels are companion phase-contrast images. Scale bars, 1 mm.
Review - Auxiliary factors: a chink in the armor of MRSA resistance to β-lactam antibiotics.
Combination agents provide an important approach to treat infectious diseases, particularly those caused by drug resistant pathogens. Indeed, applying a biologically 'rational' and systems-level paradigm to discover potent, selective, and synergistic agents would augment current (and arguably overly relied upon) empirical and serendipitous approaches to such discovery efforts. Here, we review the cellular mechanisms of β-lactam drug resistance and tolerance achieved amongst methicillin-resistant Staphylococcus aureus (MRSA) as well as their molecular targets and strategies to identify cognate inhibitors as potential combination agents to restore β-lactam efficacy against MRSA.
Roemer T, Schneider T and Pinho MG. Auxiliary factors: a chink in the armor of MRSA resistance to β-lactam antibiotics. 2013. Curr Opin Microbiol. 16:538-548. doi: 10.1016/j.mib.2013.06.012.
Absence of nucleoid occlusion effector Noc impairs formation of orthogonal FtsZ rings during Staphylococcus aureus cell division.
The Gram-positive pathogen Staphylococcus aureus divides by synthesizing the septum in three orthogonal planes over three consecutive division cycles. This process has to be tightly coordinated with chromosome segregation to avoid bisection of the nucleoid by the septum. Here we show that deletion of the nucleoid occlusion effector Noc in S. aureus results in the formation of Z-rings over the nucleoid, as well as in DNA breaks, indicating that Noc has an important role as an antiguillotine checkpoint that prevents septa from forming over the DNA. Furthermore, Noc deleted cells show multiple Z-rings, which are no longer placed in perpendicular planes. We propose that the axis of chromosome segregation has a role in determining the placement of the division septum. This is achieved via the action of Noc, which restricts the placement of the division septum to one of an infinite number of potential division planes that exist in S. aureus.
H. Veiga, A.M. Jorge and M.G. Pinho. Absence of nucleoid occlusion effector Noc impairs formation of orthogonal FtsZ rings duringStaphylococcus aureus cell division. 2011. Mol. Microbiol. 80:1366-1380. doi: 10.1111/j.1365-2958.2011.07651
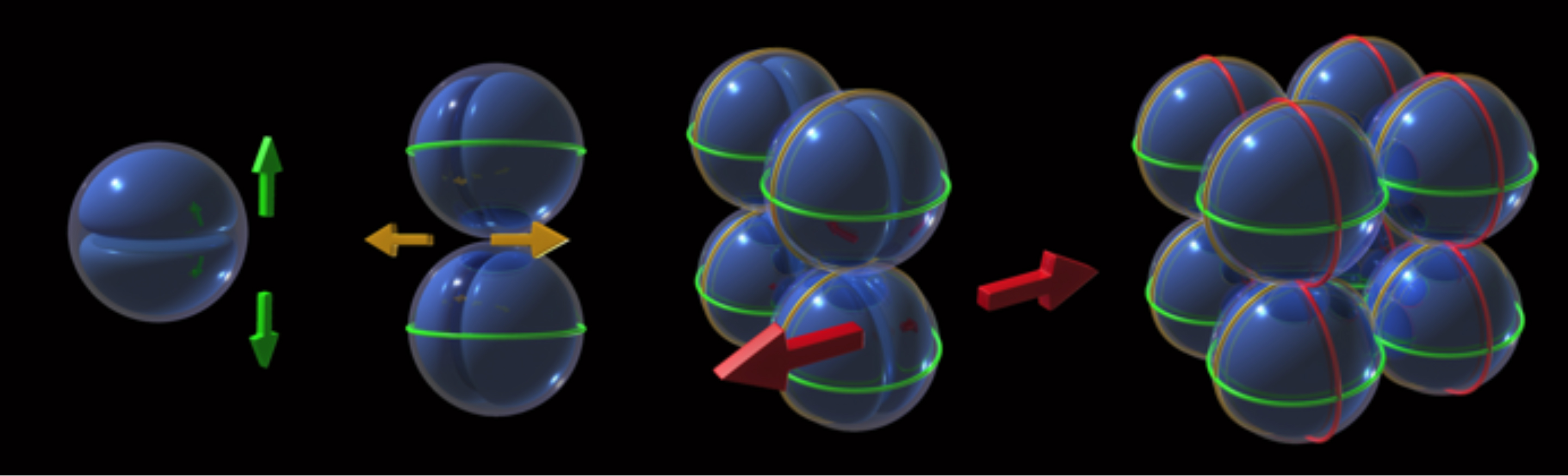
Staphylococcus aureus divides in three perpendicular planes over three consecutive division cycles
Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus
Staphylococcus aureus cell wall is characterized by an extremely high degree of crosslinking within its peptidoglycan. Penicillin Binding Protein (PBP) 4 is required for the synthesis of this highly crosslinked peptidoglycan. We found that wall teichoic acids, glycopolymers attached to the peptidoglycan and important for virulence in gram positive bacteria, act as temporal and spatial regulators of peptidoglycan metabolism, controlling the level of crosslinking by regulating PBP4 localization. PBP4 normally localizes at the division septum but in the absence of wall teichoic acids synthesis, it becomes dispersed throughout the entire cell membrane and is unable to function normally. As a consequence, the peptidoglycan of TagO null mutants, impaired in wall teichoic acids biosynthesis, has a decreased degree of crosslinking, which renders it more susceptible to the action of lysozyme, an enzyme produced by different host organisms as an initial defense against bacterial infection.
M.L. Atilano, P.M. Pereira, J. Yates, P. Reed, H. Veiga, M.G. Pinho * and S.R. Filipe*. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking inStaphylococcus aureus. 2010. Proc Natl Acad Sci U S A. 107: 18991–18996 * Both authors contributed equally to this work. doi: 10.1073/pnas.1004304107.
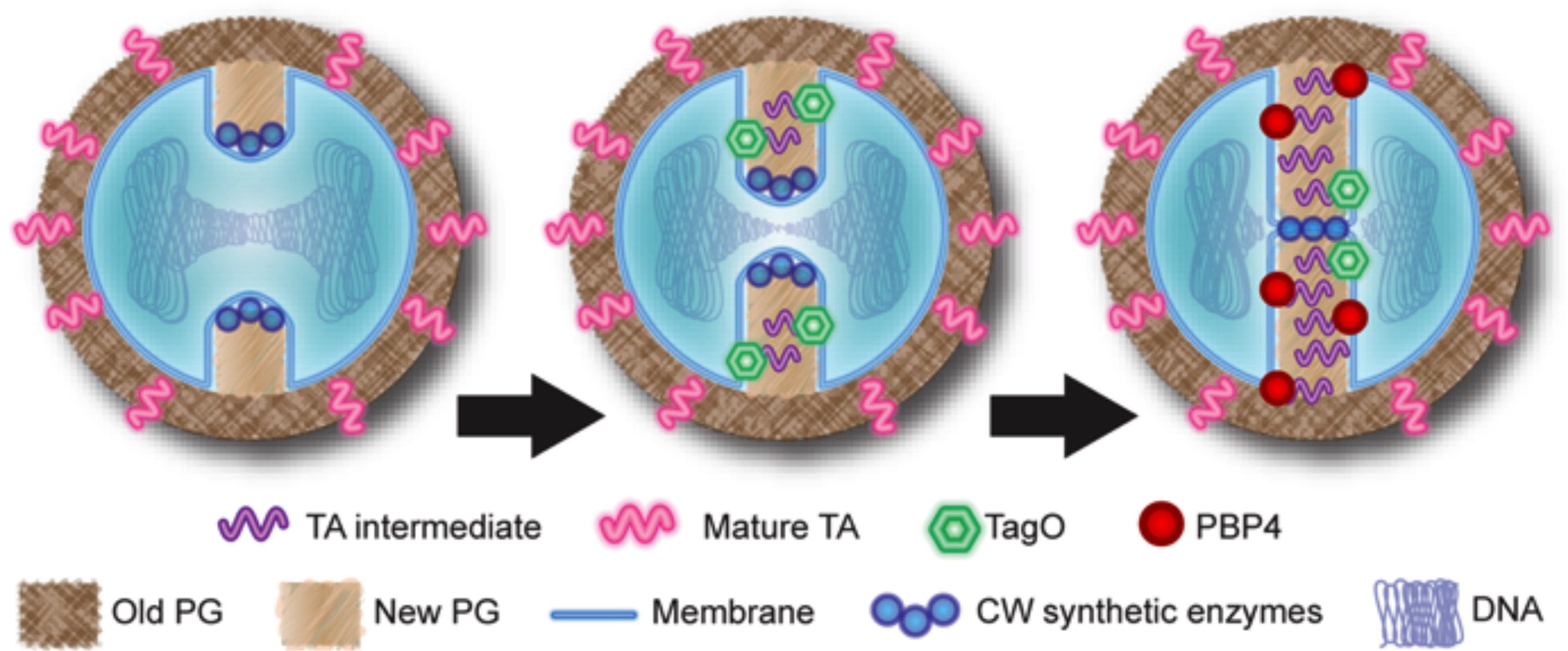 Model for the role of teichoic acids in PBP4 recruitment to the septum.
Model for the role of teichoic acids in PBP4 recruitment to the septum.
The early cell wall synthetic machinery assembles at the division site, leading to the synthesis of new peptidoglycan, with low levels of crosslinking (left). TagO (together with other wall teichoic acids synthetic enzymes) is recruited to the septum by an unknown mechanism, leading to the synthesis of intermediate molecules in wall teichoic acids biosynthesis (middle). These intermediates function as a temporal and spatial cue for PBP4 recruitment to the division septum, allowing the synthesis of highly crosslinked peptidoglycan to occur in a regulated manner (right).
Fluorescent reporters for protein cellular localization studies in Staphylococcus aureus
We have constructed a set of plasmids that allow expression, from their native chromosomal loci, of Staphylococcus aureus proteins fused to one of four different fluorescent proteins (green fluorescent protein [GFP], cyan fluorescent protein [CFP], yellow fluorescent protein [YFP], and mCherry), using two different resistance markers (kanamycin and erythromycin). We have also constructed a plasmid that allows expression of proteins from the ectopic spa locus in the S. aureus chromosome. This toolbox can be used for studies of the localization of proteins in S. aureus, a prominent pathogen in both health care and community settings.
P.M. Pereira, H. Veiga, A.M. Jorge and M.G. Pinho. Fluorescent reporters for protein cellular localization studies in Staphylococcus aureus . 2010. Appl Environ Microbiol. 76:4346–4353. doi: 10.1128/AEM.00359-10.
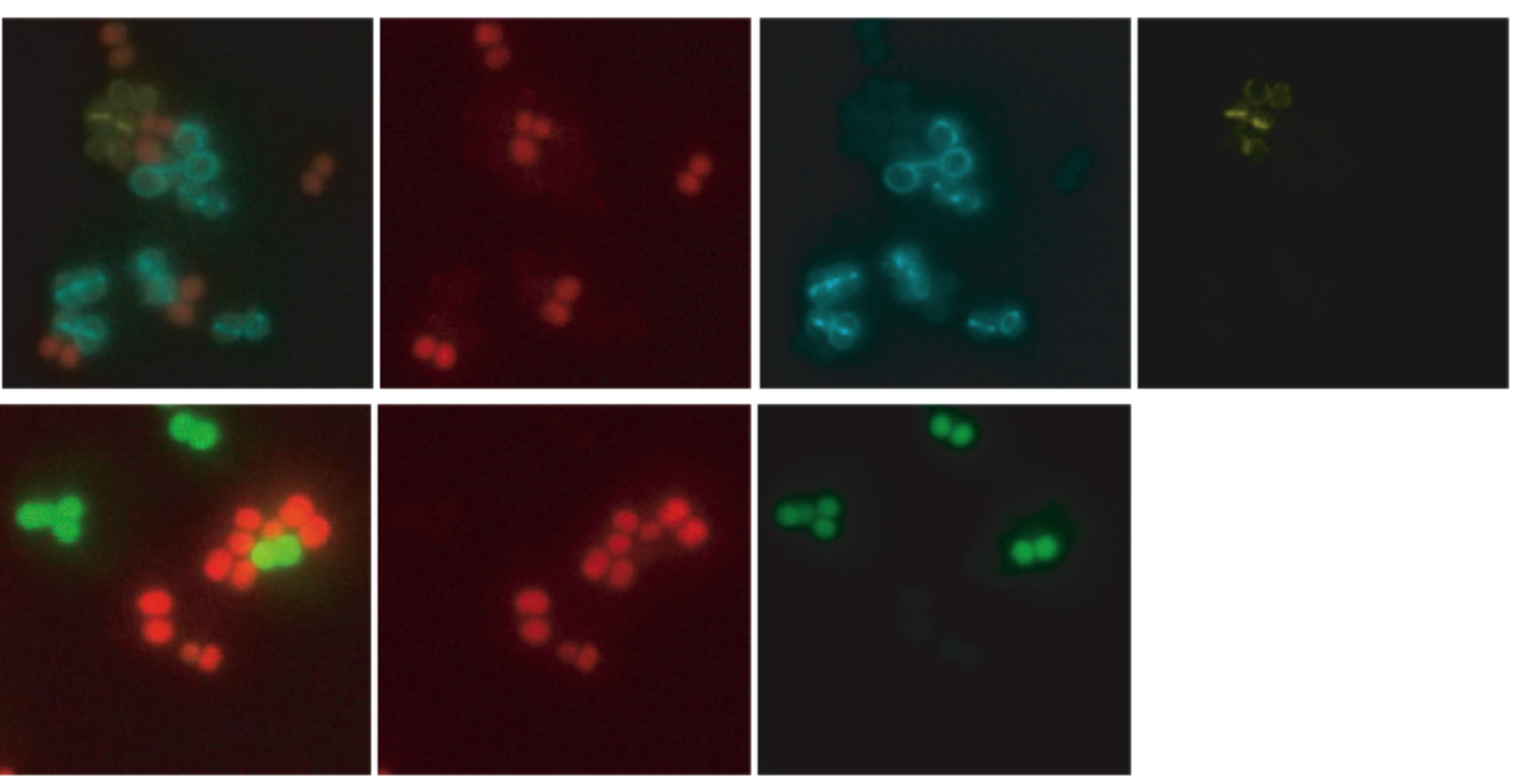
Co-visualization of S. aureus cells labeled with different fluorescent reporters . Exponential growing cultures of cells expressing different fluorescent proteins were mixed and placed on the same microscope slide. (Top) mixed culture of S. aureus strains expressing TetR-mCherry (red), EzrA-CFP (cyan ) and PBP4-YFP (yellow) fusion proteins; first image is an overlay of mCherry, CFP and YFP fluorescence. (Bottom) mixed culture of S. aureus strains expressing free GFP or TetR-mCherry in the cytoplasm; first image is an overlay of mCherry and GFP fluorescence from the same strains
Review - The different shapes of cocci.
The shape of bacteria is determined by their cell wall and can be very diverse. Even among genera with the suffix 'cocci', which are the focus of this review, different shapes exist. While staphylococci or Neisseria cells, for example, are truly round-shaped, streptococci, lactococci or enterococci have an ovoid shape (like a rugby ball). Interestingly, there seems to be a correlation between the shape of an organism and its set of penicillin-binding proteins-the enzymes that assemble the peptidoglycan, the main constituent of the cell wall. While only one peptidoglycan biosynthesis machinery seems to exist in staphylococci, two of these machineries are proposed to function in ovoid-shaped bacteria, reinforcing the intrinsic differences regarding the morphogenesis of different classes of cocci. The present review aims to integrate older ultra-structural data with recent localization studies, in order to clarify the relation between the mechanisms of cell wall synthesis and the determination of cell shape in various cocci.
Zapun, T. Vernet and M.G. Pinho. The different shapes of cocci.2008. FEMS Microbiol Rev., 32:345-360. doi: 10.1111/j.1574-6976.2007.00098.x.
Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in Vancomycin-Resistant Staphylococcus aureus
We developed a new method of fluorescence ratio imaging microscopy to compare the in vivo binding capacity and the access of a fluorescent derivative of vancomycin to the cell wall synthetic sites in isogenic pairs of vancomycin susceptible and resistant laboratory mutants and vancomycin-intermediate and susceptible clinical isolates of Staphylococcus aureus. Live cells of resistant strains were found to bind approximately 1.5 times more antibiotic but there was no correlation between the increased binding capacity and the MIC values of the strains. In both susceptible and resistant bacteria, the subcellular sites of wall synthesis were localized to the division septa but the rate of diffusion of drug molecules to these sites was reduced in resistant cells. The findings allow a reinterpretation of the mechanism of vancomycin resistance, in which the path of vancomycin to its lethal target (lipid II) is considered to be through the division septum and therefore is dependent on the stage of the staphylococcal cell cycle.
P.M. Pereira, S.R. Filipe, A. Tomasz and M.G. Pinho. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in Vancomycin-ResistantStaphylococcus aureus. 2007. Antimicrobial Agents and Chemotherapy, 51:3627-3633. doi:10.1128/AAC.00431-07
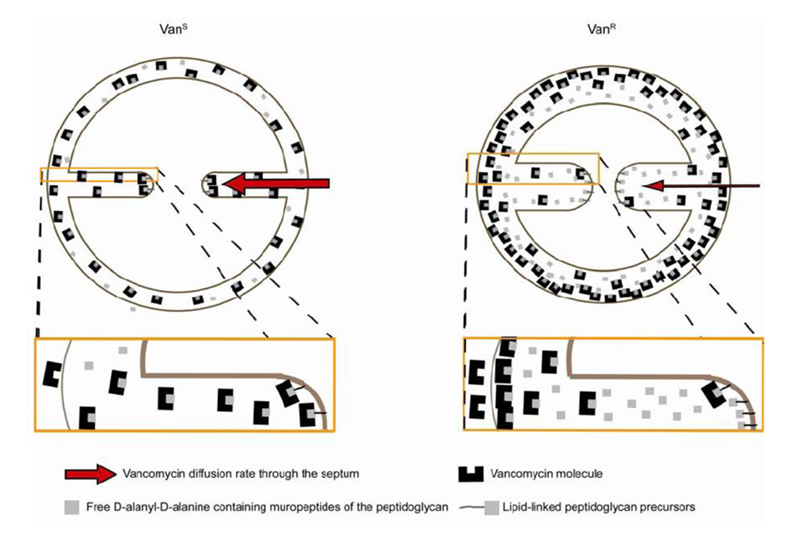
Model for vancomycin resistance in VISA strains . The path of vancomycin to its lethal target (lipid II) should be through the division septum. In resistant cells (VanR), the diffusion rate of vancomycin molecules to the septal tip is decreased, lowering the effective concentration of antibiotic that reaches the lipid-linked peptidoglycan precursor (lipid II) at the site of cell wall synthesis, per unit time, and therefore tilting the balance in favour of continued cell wall synthesis. This model implies that vancomycin efficiency may vary during the cell cycle, as the path from the outside of the cell to the lethal targets is shorter when the septum starts to be formed and longer when septum synthesis approaches completion.
Review - Bacterial cell wall synthesis - new insights from localization studies.
In order to maintain shape and withstand intracellular pressure, most bacteria are surrounded by a cell wall that consists mainly of the cross-linked polymer peptidoglycan (PG). The importance of PG for the maintenance of bacterial cell shape is underscored by the fact that, for various bacteria, several mutations affecting PG synthesis are associated with cell shape defects. In recent years, the application of fluorescence microscopy to the field of PG synthesis has led to an enormous increase in data on the relationship between cell wall synthesis and bacterial cell shape. First, a novel staining method enabled the visualization of PG precursor incorporation in live cells. Second, penicillin-binding proteins (PBPs), which mediate the final stages of PG synthesis, have been localized in various model organisms by means of immunofluorescence microscopy or green fluorescent protein fusions. In this review, we integrate the knowledge on the last stages of PG synthesis obtained in previous studies with the new data available on localization of PG synthesis and PBPs, in both rod-shaped and coccoid cells. We discuss a model in which, at least for a subset of PBPs, the presence of substrate is a major factor in determining PBP localization.
D-J. Scheffers and M.G. Pinho. 2005. Bacterial cell wall synthesis - new insights from localization studies. Microbiology and Molecular Biology Reviews, 69: 585-607.



