Biological Energy Transduction
Life depends on constant energy transduction mechanisms. In most organisms, from prokaryotes to eukaryotes the obtained energy whether from light, inorganic or organic compounds is transduced into a transmembrane difference of electrochemical potential across the prokaryotic cytoplasmatic or mitochondrial membranes. For example, non-photosynthetic organisms obtain energy by the degradation of organic food components, such as proteins, glycides and lipids, which feeds electrons to the respiratory chain. Here the electron transfer is coupled to the translocation of ions across the membrane and the energy thus released by the favourable electron transfer is transduced to the form of a transmembrane difference of electrochemical potential. This transmembrane potential is vital for solute/nutrient cell import, synthesis of ATP and motility (Figure 1). Peter Mitchell proposed the existence of such potential for the first time in his Chemiosmotic Theory.
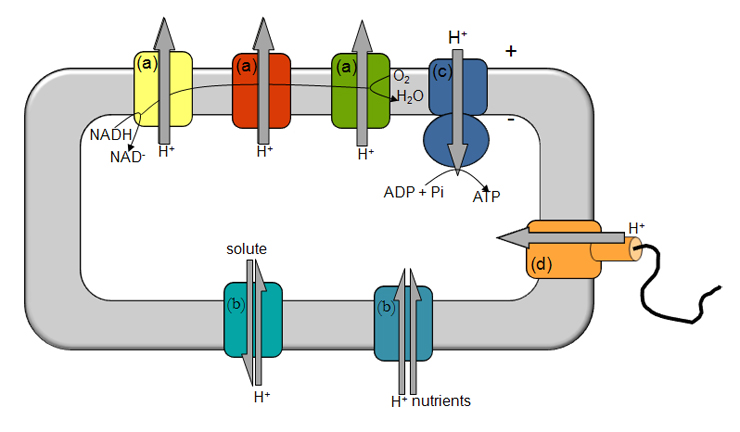
Figure 1- Illustration of energy transduction mechanisms. The electron transfer through several respiratory complexes (a) from NADH to O2 is coupled to the translocation of ions across the membrane, establishing a transmembrane difference of electrochemical potential, which is vital for solute/nutrient cell import (b), synthesis of ATP (c) and motility (d). + and – indicate the orientation of the transmembrane difference of electrochemical potential.
In our group we study the molecular mechanisms of electron transfer, ion translocation and their coupling. As model systems we use respiratory chains and their components (Figure 2). The research involves isolated proteins (wild type and recombinants) as well as reconstituted proteins or even membrane vesicles. We perform a multidisciplinary approach using a wide range of biochemical and biophysical techniques. The group is part of the Metalloproteins and Bioenergetics Unit and has several collaborations within ITQB, as well as outside the institute.

Specifically, we have been dedicated to three main research lines,
1- Complex I - CpI
Complex I catalyzes the oxidation of NADH and the reduction of quinone coupled to charge translocation across the membrane. Deficiencies on this complex have been shown to be implicated in several pathologies, namely neurodegenerative diseases such as Leber’s hereditary optic neuropathy, Parkinson and Dystonia disorders. The study of this enzyme is currently a hot-topic in the bioenergetics’ field, with a recent gathering of structural and functional data, including its X-ray structure.
We have been studying complex I using Rhodothermus marinus as a model system, namely investigating its electron transfer kinetics and quinone reduction, H+ and Na+ translocation and the coupling mechanism of electron transfer to ion transport. We have recently extended our approach to complex I from Escherichia coli and Paracoccus denitrificans, the most studied prokaryotic complexes I.
2- Alternative Complex III - ACIII
Despite the large diversity and flexibility observed in the prokaryotic electron transfer chains, the cytochrome bc1 complex family was thought to be the only able to perform quinol: electron acceptor oxidoreductase activity. However, in our group we have identified for the first time an alternative complex III, ACIII, which is structurally unrelated to the former. We observed a relation of the ACIII with members of the complex iron-sulfur molybdoenzyme family and we concluded that ACIII is a new complex composed by a novel combination of modules already identified in other respiratory complexes.
3- Heme-copper oxygen reductases - HCO
Heme-copper Oxygen reductases (HCOs) are the main enzymes responsible for reduction of oxygen to water in respiratory chains. These membrane-bound enzymes are present in the three life domains, Bacteria, Archaea and Eukarya, catalyzing the last reaction of aerobic respiratory chains. We have proposed a classification scheme based on the fingerprint of the proton conducting channels of these enzymes, and thus HCO were divided into three types: A (further divided into A1 and A2), B and C Type. We have been performing a systematic and comparative functional and spectroscopic study of representative enzymes from the three types aiming at identifying the possible common denominator in haem-copper oxygen reductases and thus revealing the key elements responsible for oxygen reduction, including the uptake of the substrate protons, for the proton pumping mechanism and for the coupling of these two events.



