PTMs in Proteome Regulation

We are at a standstill in what comes to crop yield production and the demand for yield increase is adamant, mostly due to the population growth. Adding to this, there is the obvious climate unpredictability that is affecting regular crop production.
To address these questions, scientists are searching for new ways to improve C- fixation/photosynthesis and to increase plant tolerance to stress. We are engaged in this effort by exploring the regulation of key proteins by post-translational modifications (PTMs). There is an obvious lack of knowledge in plants regarding how PTMs regulate the proteome, as compared to what is known in the animal field. Because PTMs can act in the cell proteome to rapidly change its properties, our hypothesis is that by “manipulating” these mechanisms, we may prepare the cell for a first line of response (rapid response) to environmental changes. This novel approach will hopefully contribute with new plant improvement strategies.
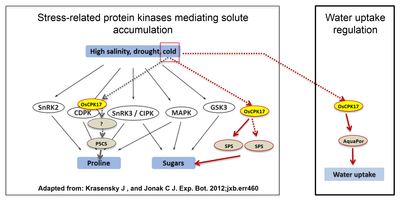
In general, stress and metabolic signaling networks interact allowing plants to respond to biotic (herbivory and pathogen attack) and abiotic (drought, cold, heat and osmotic stresses including salinity) stresses. At the interface between these two major signaling systems are, among others, calcium-dependent protein kinases (CPKs). These are dual-action enzymes, capable of sensing the calcium signal arising from stress and transmitting the encoded information trough phosphorylation of downstream targets. In rice, CPKs are a family of 31 genes, and we have previously shown the involvement of several of them in both biotic and abiotic stress response. Particularly, OsCPK17 was shown to be responsible for the phosphorylation of key enzymes in rice anabolic processes. We are using state-of-the-art mass spectrometry methodologies to identify and quantify in vivo targets of rice kinases, combining this approach with biochemical and molecular biology approaches to study the functional relevance of those findings.
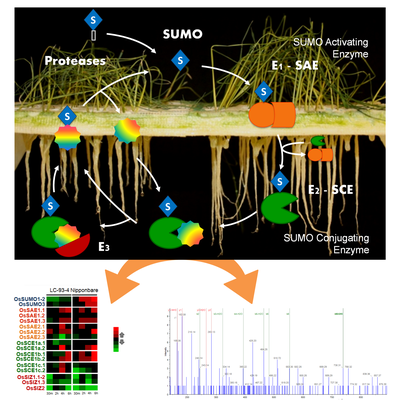 SUMOylation is an essential post-translational modification that affects several cellular processes, from gene replication to stress response. Studies using the SUMO (de)conjugation machinery have provided evidence regarding its potential to improve crop performance and productivity under normal and adverse conditions. Our results show that the transcriptional response of the components of the rice SUMOylation machinery is stress- and genotype-dependent, when comparing stress tolerant and sensitive rice genotypes. Rice transformed lines over-expressing some of these components in panicles were obtained and show positive impact in yield. We are studying the role of these components by characterizing their function in development and stress tolerance acquisition, using single/double-KO mutants and transgenic rice plants.
SUMOylation is an essential post-translational modification that affects several cellular processes, from gene replication to stress response. Studies using the SUMO (de)conjugation machinery have provided evidence regarding its potential to improve crop performance and productivity under normal and adverse conditions. Our results show that the transcriptional response of the components of the rice SUMOylation machinery is stress- and genotype-dependent, when comparing stress tolerant and sensitive rice genotypes. Rice transformed lines over-expressing some of these components in panicles were obtained and show positive impact in yield. We are studying the role of these components by characterizing their function in development and stress tolerance acquisition, using single/double-KO mutants and transgenic rice plants.



