Genetics, Biochemical and Evolutionary Mechanisms in Staphylococci
1. Antibiotic Resistance Mechanisms
Staphylococcus aureus is a challenge for antimicrobial chemotherapy. In particular methicillin resistant S. aureus (MRSA) create severe problems, because in addition to resistance to β-lactams, these bacteria are extremely resourceful in accumulating and developing resistance to other classes of antibiotics.
Role of peptidoglycan synthesis pathway in the resistance mechanisms
Ana Madalena Ludovice, Rita Sobral, Teresa Figueiredo, Inês Grilo, and Raquel Portela.
Peptidoglycan, a major component of the cell wall of bacteria, is a unique and essential polymer and its biosynthetic pathway is the target of the most powerful antibacterial agents developed. We are currently exploring some unknown aspects of the cell wall biosynthesis in S. aureus.
A. Characterization of the amidation step of the stem peptide of S. aureus peptidoglycan
The murT-gatD operon was shown to encode the proteins responsible for the amidation of peptidoglycan in S. aureus. The lack of amidation resulted in a decrease in oxacillin and in lysozyme resistance, implying that MurT and GatD are involved not only in antibiotic resistance but also in the virulence potential of the strain. These results were recently published (Figueiredo et al. 2012. PLOS pathogens. doi:10.1371/journal.ppat.1002508) and are included in the PhD Thesis work of T. Figueiredo. The work was performed in collaboration with A. Tomasz (The Rockefeller University, USA) and W. Vollmer (Newcastle University, UK).
| All isolates were identified by phenotypic and molecular methods. Isolates involved in ovine or bovine mastitis are being typed by the several molecular methodologies used in our laboratory. Typing of these isolates should allow the identification of clones circulating in each herd, which may contribute to the understanding of the spread of the disease. By comparing these data with results from other laboratories we also aim to address the relatedness of these isolates with those implied in episodes of mastitis in other countries. Additional characterization of these isolates includes screening for virulence and antibiotic resistance genes. We are currently studying the impact of the impairment of murT-gatD operon in CA-MRSA (Community Acquired strains). The 3-D structure of the MurT-GatD complex is being determined in collaboration with J. Trincão (University of Bath, UK). The two proteins present new domains and domain arrangements which have not yet been described. The interaction between MurT and GatD is being analyzed through NMR analysis in collaboration with E. Cabrita (Requinte, FCT, UNL). |
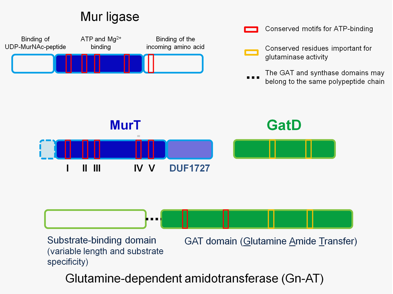
Figure 1. Protein regions necessary for the cooperative function of MurT and GatD proteins. The top panel represents the general topology of Mur ligase family proteins, with three domains. The only domain showing homology with MurT is the central domain involved in nucleotide binding. The lower panel represents the modular structure of Gn-ATs, with a synthase domain and a GAT domain which has glutaminase activity motifs and ATP-binding motifs. |
B. Implication of the two-component system VraSR on intermediate resistance to vancomycin in S. aureus
The emergence of multi-drug resistant S. aureus strains exhibiting intermediate and high resistance to vancomycin (VISA and VRSA, respectively) and their global dissemination is a serious concern in clinical practice. Besides vancomycin, very few antibiotics remain effective against staphylococcal infections. The mechanism of intermediate resistance to vancomycin at both genetic and biochemical levels is currently being investigated in our laboratory. The first Portuguese VISA strain showing homogeneous resistance to vancomycin (MIC of 4 µg/ml) was isolated in 2006 from a wound of a female patient hospitalized at the orthopedics ward of Hospital de São Marcos-Braga located in the North of Portugal (Gardete, S. et al., 2008).
Recent studies suggest that structural changes in the two-component system VraSR - a key regulatory element in the adaptative response of S. aureus to cell wall active agents - and stimulation of the VraSR regulon appear to be part of the strategy used by this bacterium to overcome the deleterious effect of vancomycin. Understanding the mode of action VraSR and better defining its regulon should identify new suitable targets for novel antibacterial compounds agents against VISA strains.
C. Genome comparison of a S. aureus mutant showing decreased antibiotic resistance and its revertant strain
Through whole genome sequencing, we detected genome changes between the parental strain COL, a murF mutant with decreased oxacillin resistance and a stable revertant F9H2, selected for growth at high oxacillin concentrations.
A single nucleotide change was detected in F9H2 genome: a missense mutation in the α chain of phenylalanyl-tRNA synthetase which catalyzes the conversion of phenylalanine and ATP into the stable intermediate phenyl-adenylate. We are now constructing a conditional mutant for the tRNAPHE synthetase gene to determine if this single mutation is responsible for reestablishing the high and homogeneous resistance phenotype.
This work is being performed in collaboration with A. Tomasz and is part of the work plan of student R. Portela.
D. Functional study of the cytoplasmic steps of peptidoglycan synthesis
The cell wall biosynthetic steps in S. aureus have been the subject of intense research, mainly focusing on implications to methicillin resistance and on the physiology of the cell (Sobral, R.G. et al., 2006; Sobral, R.G. et al., 2007). Transcriptomic studies are being followed to clarify the genetic networks involved in the cell wall metabolism and are part of the work plan of the Research student R. Portela.
Selected publications
- Kim, C., C. Milheiriço, S. Gardete, M. A. Holmes, M. T. G. Holden, H. de Lencastre, and A. Tomasz. 2012. Properties of a novel PBP2A protein homologue from Staphylococcus aureus strain LGA251 and its contribution to the ß-lactam resistant phenotype. J. Biol. Chem. jbc.M112.395962. First Published on September 12, 2012, doi:10.1074/jbc.M112.395962
- Arêde, P., C. Milheiriço , H. de Lencastre, and D. C. Oliveira. 2012. The anti-repressor MecR2 promotes the proteolysis of the mecA repressor and enables optimal expression of β-lactam resistance in MRSA. PLoS Pathogens 8:e1002816.
- Figueiredo, T.A., R. G. Sobral, A. M. Ludovice, J. M. F. de Almeida, N. K. Bui, W. Vollmer, H. de Lencastre and A. Tomasz. 2012. Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus. PLoS Pathogens 8:e1002508.
- Oliveira, D.C., and H. de Lencastre. 2011. Methicillin-resistance in Staphylococcus aureus is not affected by the overexpression in trans of the mecA gene repressor: a surprising observation. PLoS One 6:e23287.
- Milheiriço C., A. Portelinha, L. Krippahl, H. de Lencastre, and D. C. Oliveira. 2011. Evidence for a purifying selection acting on the beta-lactamase locus in epidemic clones of methicillin-resistant Staphylococcus aureus. BMC Microbiology 11:76.