Prediction of pKa's and redox potentials
Thermodynamics of Electron-Proton Coupling
In a solution with given pH and electrostatic (electrode) potential E, a protein molecule undergoes a continual exchange of titrating electrons and protons between its sites and the solution. This molecular aspect of the joint binding equilibrium of electrons and protons is reflected on the properties of protein molecules, having both energetic and entropic consequences [1]. Understanding these effects is thus determinant for the study of the thermodynamic aspects of electron-proton coupling in protein molecules [1].
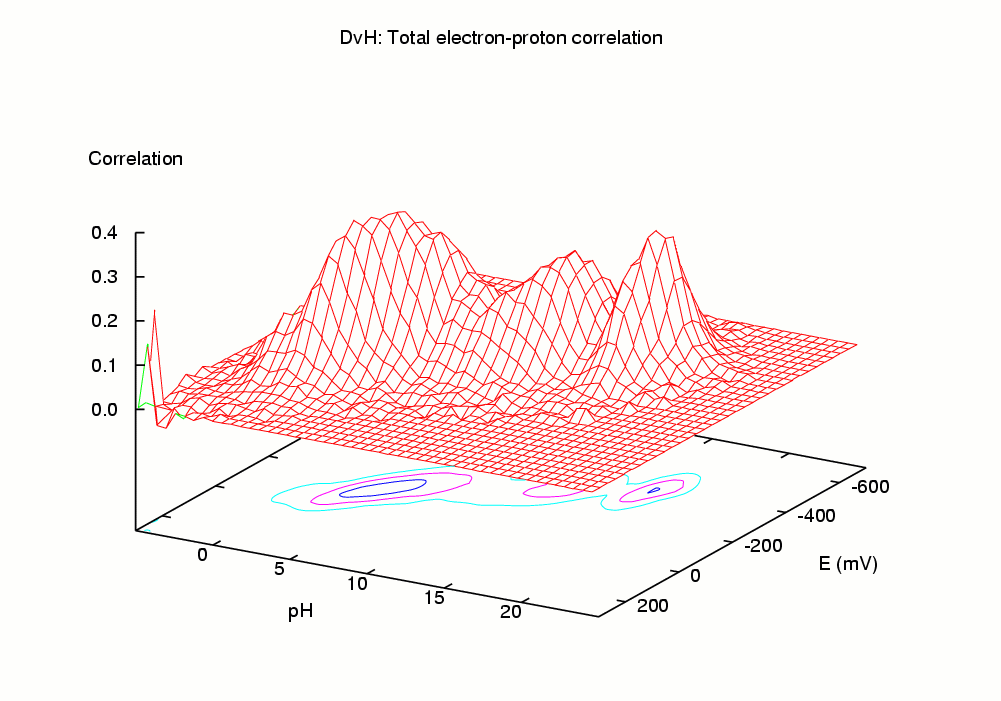
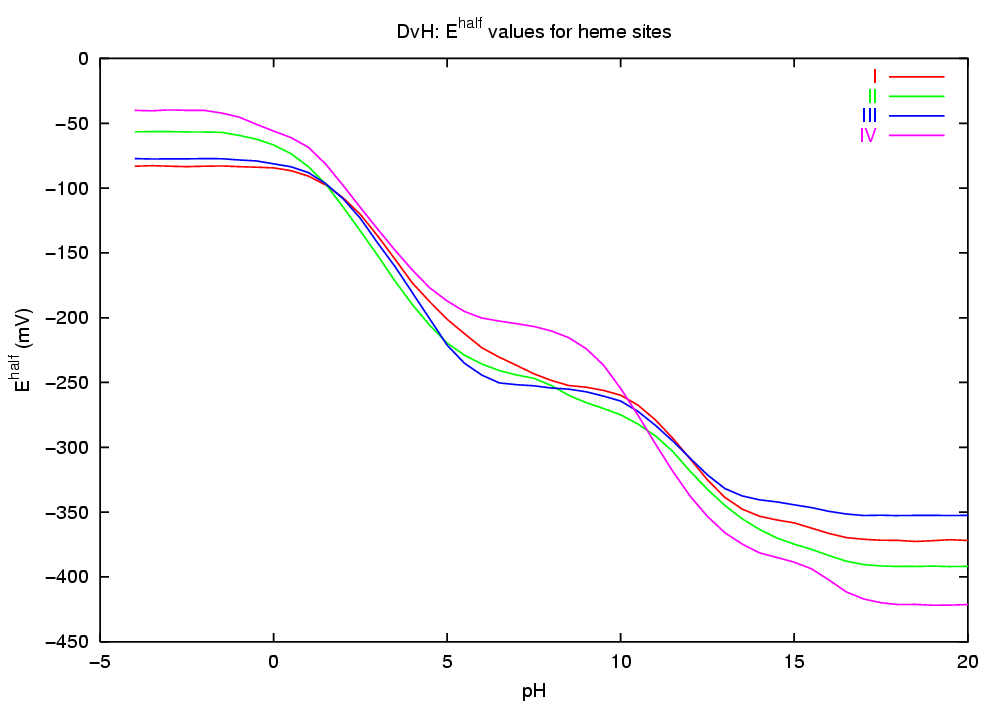
The simulation of this microscopic "dynamic" equilibrium can be achieved by the theoretical and computational tools we have recently developed [1-4]. The programs developed perform a Monte Carlo simulation of the joint equilibrium of electrons and protons, for a range of pH vs E, using as input a set of intrinsic affinities (pKint and Eint values) and a set of pairwise interactions. Among the properties computed are average (macroscopic) occupancies, site-site correlations, occupational entropies, etc. In particular, the correlation between a pair of binding sites provides a measure of coupling (or linkage) between the sites which captures both direct and indirect effects [1]. The pH-dependence of midpoint potentials (redox-Bohr effect) can also be studied.
Tautomerism in pKa Calculations
Continuum electrostatic models [5-6] can treat protons in three different ways: the first approach assigns an average charge to the protonated form of the protonatable sites not accounting for the alternative proton positions in the protonatable sites; the second approach tries to deal with this aspect by selecting, for each site, one of the proton positions as the one appropriate to the protonated form; and the third one considers all proton isomers, also called tautomers. The inclusion of proton tautomerism in protonation equilibrium calculations models the reorganization of the protein to the protonation changes themselves considering the protonation-conformation coupling [2].
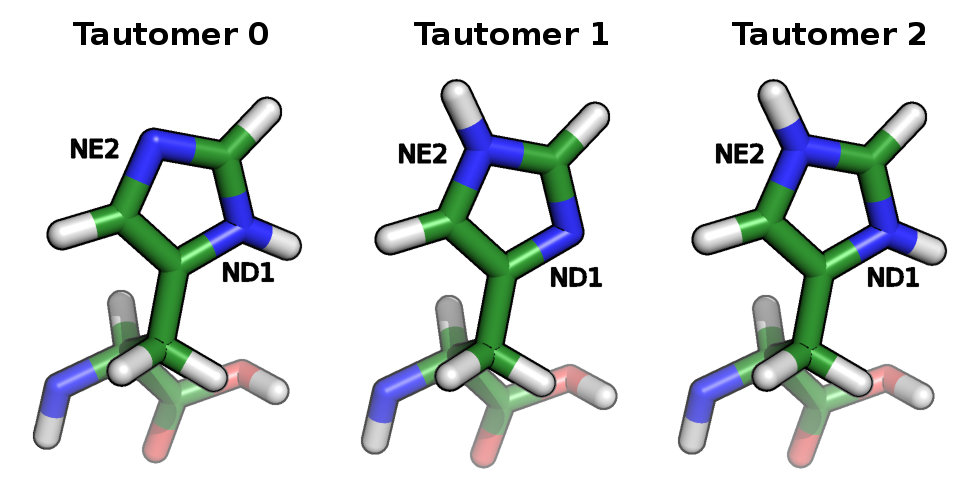
Histidine tautomers
Prediction of pKa’s using Linear Response Approximation
The linear response approximation (LRA) using two end points has been successfully used to determine pKa values in proteins [7-9]. LRA can be used to compute the equilibrium pKa (pKaeq) of the site of interest, which is given by:
pKaeq = ½[⟨pKaeq(c)⟩P + ⟨pKaeq(c)⟩U]
where pKaeq(c) is the pKa if the protein were fixed in conformation c, and the angle brackets denote averages over the peptide conformations sampled from the MD runs with the site protonated (P) or unprotonated (U). It should be noted that the pKaeq is a pH-dependent quantity referring to the dissociation equilibrium constant; the midpoint pKa, corresponding to the pH value of half-protonation, is obtained afterward as the pH value for which pKaeq = pH.
References
[1] Baptista, A.M., Martel, P.J., Soares, C.M. (1999) Simulation of electron-proton coupling with a Monte Carlo method: Application to cytochrome c3 using continuum electrostatics. Biophys. J., 76:2978-2998.
[2] Baptista, A.M., Soares, C.M. (2001) Some theoretical and computational aspects of the inclusion of proton isomerism in the protonation equilibrium of proteins. J. Phys. Chem. B, 105:293-309.
[3] Teixeira, V.H., Soares, C.M., Baptista, A.M. (2002) Studies of the reduction and protonation behavior of tetraheme cytochromes using atomic detail. J. Biol. Inorg. Chem., 7:200-216.
[4] Teixeira, V. H., Cunha, C. A., Machuqueiro, M., Oliveira, A. S. F., Victor, B. L., Soares, C. M., Baptista, A. M. (2005) On the use of different dielectric constants for computing individual and pairwise terms in Poisson-Boltzmann studies of protein ionization equilibrium. J. Phys. Chem. B 109:14691-14706.
[5] Sharp, K. A., Honig, B. (1990) Electrostatic interactions in molecules: Theory and Applications. Annu. Rev. Biophys. Chem., 19:301-332.
[6] Honig, B., Nicholls, A. (1995) Classical Electrostatics in Biology and Chemistry. Science, 268:1144-1149.
[7] Lee, F. S., Chu, Z.-T., Bolger, M. B., Warshel, A. (1992) Calculations of antibody-antigen interactions: microscopic and semi-microscopic evaluation of the free energies of binding of phosphorylcholine analogs to McPC603. Protein Eng., 5:215-228.
[8] Eberini, I., Baptista, A. M., Gianazza, E., Fraternali, F., Beringhelli,T. (2004) Reorganization in Apo- and Holo-ß-Lactoglobulin Upon Protonation of Glu89: Molecular Dynamics and pKa Calculations. Proteins, 54:744-758.
[9] Machuqueiro, M., Campos, S. R. R., Soares, C. M., Baptista, A. M. (2010) Membrane-Induced Conformational Changes of Kyotorphin Revealed by Molecular Dynamics Simulations. J. Phys. Chem. B, 114:11659–11667.



