Coagulase-negative staphylococci
1. Molecular epidemiology and evolution of methicillin-resistant Staphylococcus epidermidis (MRSE)
Joana Rolo, Maria Miragaia
A. Hospital-associated methicillin-resistant S. epidermidis (HA-MRSE)
In the last decades Staphylococcus epidermidis has emerged as one of the most significant nosocomial pathogens, mainly associated to indwelling catheters and implanted devices. Nowadays, approximately 60-70% of the S. epidermidis strains isolated in the hospital environment are resistant to methicillin and to virtually all antimicrobial classes.
Studies conducted in our laboratory, revealed key features of S. epidermidis epidemiology, evolution and population structure, namely the existence of S. epidermidis epidemic clones that evolved quickly through recombination and frequent exchange of genetic mobile elements. Of special relevance, we found that S. epidermidis strains acquired the mobile genetic element carrying the determinant of methicillin resistance, SCCmec, a high number of times. In addition, our data indicates that S. epidermidis may be functioning as a factory of new SCCmec types and non-mec SCC that eventually can be transferred to other staphylococcal species, functioning as an important reservoir of antimicrobial resistance and virulence genes.
In the light of these new findings on S. epidermidis epidemiology, we reviewed the usefulness and applicability of pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and SCCmec typing to characterize S. epidermidis strains. Using a quantitative assessment we found that the combination of the PFGE and SCCmec typing results was suitable to define a S. epidermidis clone.
Our present research focus on the application of whole genomic approaches to understand the evolution of nosocomial S. epidermidis epidemic clones.
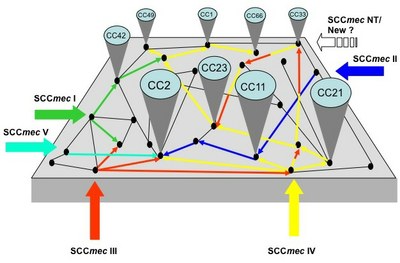 |
Figure 1: Model proposed for the emergence and evolution of nosocomial MRSE. Source: Miragaia M; The evolution of methicillin-resistantStaphylococcus epidermidis: diversity in genetic backgrounds and in the structure of the resistance determinant SCCmec. |
B. Community-associated methicillin-resistant S. epidermidis (CA-MRSE)
The most common SCCmec type found in MRSE is SCCmec type IV, which is 99% identical to that found in SCCmec IV from MRSA. This, associated to the fact that SCCmec type IV confers no fitness cost to bacteria that carry it, led to the hypothesis that MRSE were the donors of SCCmec type IV to MSSA in the community leading to the emergence of CA-MRSA. To test this hypothesis S. epidermidis were collected from healthy persons in the community and characterized by appropriate typing methods.
Contrarily to what was expected the rate of community-associated methicillin-resistant S. epidermidis CA-MRSE was found to be low, suggesting that CA-MRSE are not reservoirs of SCCmec IV for CA-MRSA.
Additionally, we observed that the most common S. epidermidis clones in the hospital coincided with those found in the community, revealing the lack of community-hospital barriers in the dissemination of S. epidermidis. More importantly, we found that adaptation to hospital environment was associated to the acquisition, diversification and amplification of the determinant of methicillin resistance, SCCmec.
Presently we are interesting in studying the factors promoting SCCmec acquisition and diversification in this species.
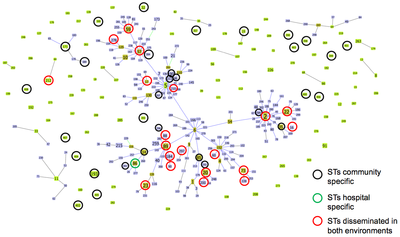 |
| Figure 2: Analysis of MLST S. epidermidis data by goeBURST algorithm; circles of different colors indicate STs that were found in the hospital and community of both environments. Source: Rolo et al. JAC. 2012. |
C. Molecular characterization of farm-associated methicillin resistant S. epidermidis (FA-MRSE)
Most of the studies on the epidemiology of MRSE focused on isolates of human origin. Much less is known regarding the epidemiology of S. epidermidis of animal origin. In order to define the population structure of S. epidermidis among animals we are presently characterizing a collection of methicillin-resistant and methicillin-susceptible S. epidermidis (MSSE) recovered from bovine and ovine by a combination of molecular typing techniques.
2. Molecular characterization of methicillin-resistant coagulase-negative staphylococci other than S. epidermidis
Ons Bouchami, Maria Miragaia
In spite of the currently recognized clinical relevance of coagulase-negative staphylococci, for decades coagulase-negative staphylococci were not identified at species level and were considered as a homogeneous group of bacteria with similar phenotypic and genotypic characteristics, what hindered the understanding of its epidemiology.
The detailed molecular characterization performed by our group of each coagulase-negative species in separate showed for the first time the singular epidemiology of S. epidermidis, Staphylococcus haemolyticus and Staphylococcus hominis, in terms of clonality, genetic diversity and reservoirs of SCC elements.
Presently, we are performing studies that aim to describe epidemic clones of S. hominis and S. haemolyticus, through the molecular characterization of isolates from different geographic regions using molecular typing techniques.
Moreover, we intend to extend these studies to other clinically relevant Staphylococcus species in the hospital and veterinary settings.
We expect that the understanding of the specific characteristic of the epidemiology of the different coagulase-negative Staphylococcus species will help to design informed infection control strategies adapted to the individuality of each species. Moreover, these studies might provide insights into the role of each species in the evolution of the determinant of methicillin resistance, SCCmec.
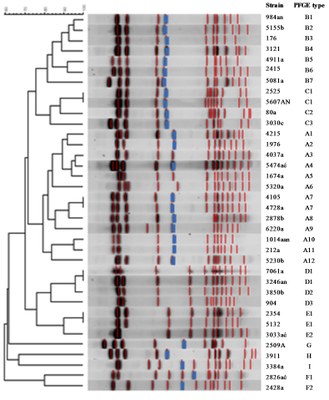 |
| Figure 3: Dendrogram of SmaI PFGE macrorestriction patterns of methicillin-resistant S. haemolyticus isolates. The SmaI-mecA hybridization band is depicted in blue. Source: Bouchami et al. EJCMID. 2012. |
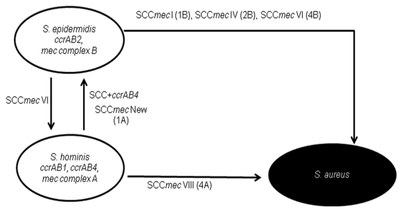 |
| Figure 4: Model for the contribution of S. hominis species for SCCmec evolution. Source: Bouchami et al. PLoS ONE. 2011. |
3. Development of new molecular typing methods to characterize CoNS
Catarina Milheiriço, Nuno Faria, Ons Bouchami, Joana Rolo, Celine Coelho, Maria Miragaia
To characterize the molecular epidemiology of each CoNS species the use of reliable methods for identification of Staphylococcus is fundamental. Phenotypic methods usually provide ambiguous identification and molecular methods are considered to be the most trustworthy. We have previously developed a PCR-based typing method for Staphylococcus species identification, based on the amplification of the internally transcribed spacer between 16S-23S. However, this method cannot differentiate at the sub-species level.
In order to overcome this drawback, we are currently involved in the validation of a sequencing-based method to identify Staphylococcus at the species and subspecies level.
Molecular typing tools described for S. aureus cannot be applied directly to CoNS what requires the development of new typing methods to study these species. We have recently contributed to the establishment of the multilocus sequence typing scheme for S. epidermidis and are presently involved in the development of the MLST scheme for S. hominis (in collaboration with Ashley Robinson, University of Mississippi Medical Center, Jackson, USA).
Moreover, we are currently validating a sequenced-based method to type SCCmec type V, targeting ccrC (see the new typing methods in S. aureus item). This method will be particularly useful to type S. haemolyticus isolates in which SCCmec type V is the most common cassette.
Selected references
Gordon R.J., M. Miragaia, A.D. Weinberg, C. J. Lee, J. Rolo, J. C. Giacalone, M. S. Slaughter, P. Pappas, Y. Naka, A. J. Tector, H. de Lencastre, and FD Lowy. 2012. Staphylococcus epidermidis colonization is highly clonal Across US Cardiac Centers. 2012. J Infect Dis. 205(9): 1391-1398.
Bouchami, O., A. Ben Hassen, H. de Lencastre, and M. Miragaia. 2012. High prevalence of mec complex C and ccrC is independent of SCCmec type V in S. haemolyticus. European Journal of Clinical Microbiology and Infectious Diseases. 31(4):605-14.
Rolo, J., H. de Lencastre, and M. Miragaia. 2012. Strategies of adaptation of Staphylococcus epidermidis to hospital and community: amplification and diversification of SCCmec. J. Antimicrob. Chemother. 67:1333-41.
Bouchami, O., A. Ben Hassen, H. de Lencastre, and M. Miragaia. 2011. Molecular epidemiology of methicillin-resistant Staphylococcus hominis (MRSHo): low clonality and reservoirs of SCCmec structural elements. PLoS ONE. 10.1371/journal.pone.0021940.
Miragaia, M., H. de Lencastre, H. F. Chambers, F. Perdreau-Remington, J. Lin, K. I. Wong, K. A. King, G. F. Sensabaugh, M. Otto, and B. A. Diep. 2009. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLOSone: 4(11):e7722.
Miragaia, M., J. A. Carriço, J. C. Thomas, I. Couto, M. C. Enright, and H. de Lencastre. 2008. Comparison of molecular typing methods for characterization of Staphylococcus epidermidis: proposal for clone definition. J. Clin. Microbiol. 46: 118-29.
Miragaia, M., J. C. Thomas, I. Couto, M. C. Enright, and H. de Lencastre. 2007. Inferring a population structure for Staphylococcus epidermidis using multilocus sequence typing (MLST) data. J. Bacteriol. 189:2540-52.



