Cell Division Dynamics Unveiled
Oeiras, 14 March 2024
Bacterial cell division is key for bacteria to proliferate. This complex process is an interesting, yet underexplored, target for new antibiotics. Over the last years, the Bacterial Cell Biology lab, led by Mariana Gomes de Pinho, at ITQB NOVA, has been working to understand the molecular details that organize cell division, funded by three consecutive ERC grants. The researchers use Staphylococcus aureus, an important pathogen which is currently the second cause of death by antibiotic resistant infections worldwide, as a model organism.
At the heart of bacterial cell division is the protein FtsZ, a homologue of the protein tubulin, essential to construct the divisome – a complex assembly of proteins that drives cell division. FtsZ undergoes treadmilling, a process in which its filaments continuously grow on one end and shrink on the other, leading to movement of the filaments around the division site. Until now, FtsZ treadmilling was thought to drive the movement of peptidoglycan synthases, the proteins that make peptidoglycan, around the division site.
This idea was overturned when Simon Schäper, the first author of a study published today in Nature Microbiology, implemented new cutting-edge single-molecule imaging techniques to follow the movement of individual proteins in S. aureus cells. “I was able to observe the directional movement around the division site of three proteins involved in peptidoglycan synthesis: FtsW, PBP1 and DivIB”, the researcher explains. Surprisingly, this movement was driven by the synthesis of a cell wall component, the septal peptidoglycan, rather than by FtsZ treadmilling. “We found that antibiotics that target peptidoglycan synthesis, such as beta-lactams, are able to stop that movement”, Schäper says.
“Historically, we started by learning what is the function of proteins, then how they assemble into functional multi-protein complexes”, says Mariana Pinho. “More recently, in the last two decades, advances of fluorescence microscopy allowed us to determine where and when proteins and complexes localize inside bacterial cells. Now, the ability to visualize single molecules in live cells allows us to go one step further and study the dynamics of proteins and complexes”. This brings research one step closer to understanding the organization of bacterial cells and how different antibiotics affect this organization.
This work was done in the framework of an ERC Consolidator grant awarded to Mariana Pinho in 2018, to identify new regulators of the bacterial cell cycle and develop new tools for antibiotic research.
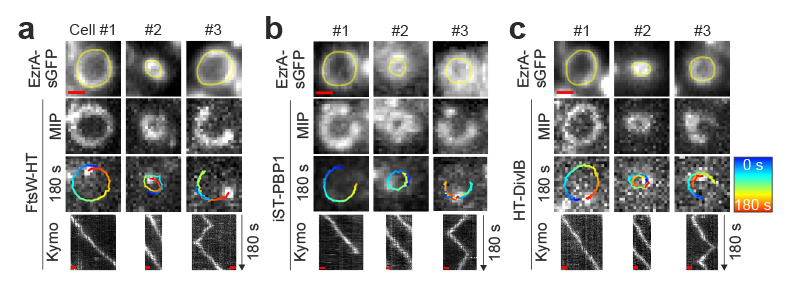
Cell constriction requires processive septal peptidoglycan synthase movement independent of FtsZ treadmilling in Staphylococcus aureus
Schaeper S, Brito AD, Saraiva BM, Squyres GR, Holmes MJ, Garner EC, Hensel Z, Henriques R, Pinho MG., Nature Microbiology







