Detected the structural points that allow SARS-CoV-2 to deceive the immune system
A recent study, published in the Journal PLOS Pathogens, focuses on the structure of the Spike protein, which is essential for the virus to enter cells and the primary target of currently administered vaccines. Knowing and being able to predict how these changes affect the course of infection is an important weapon when adjusting strategies to control the pandemic of COVID-19.
By mid-April 2021, there were more than 140 million cases of COVID-19 worldwide. High transmission inevitably increases the likelihood of new variants evolving. These include those variants that give the SARS-CoV-2 virus the ability to avoid the immune system, escaping the host without compromising its replication. "We know that the virus is changing, and it is important that we can understand and predict which mutations affect the course of the infection. In this study, we were able to show that there are two points on the Spike protein that are prone to change, to invade the immune system. This kind of knowledge is crucial for us to be able to anticipate the virus and adjust our strategies to fight the pandemic," describes Maria João Amorim, a senior researcher at Instituto Gulbenkian de Ciência (IGC) who led the study.
The study developed at the IGC in collaboration with teams led by Cláudio M. Soares, from ITQB NOVA, and Helena Soares, from CEDOC, focused on the structure of the Spike protein of SARS-CoV-2. To determine the effect of changes in this protein, the research team used techniques that allow them to express it in viral particles that are not harmful, that are easy to study, and that do not require high-security laboratories. By adding antibodies, produced after infection or vaccination, to cells and viral particles in culture, its possible to measure the protection they exert against each variant. "Using this technique, we have detected two mutations at different points in Spike, that cause the virus to evade antibodies generated after infection or vaccine administration. In addition, this occurs without compromising its entry into cells, because it does not affect the binding to the cell receptor necessary for this process. With these changes, the virus continues to enter the cells without great cost for the infection and avoids the recognition by the antibodies", explains Maria João Amorim.
Diana Lousa, a researcher at ITQB NOVA, recalls how they started from a simple idea: "What if we looked simultaneously at the data from the connection to the antibodies and our receptor? Through molecular simulation and analysis of dozens of structures, it was possible to predict which mutations could give the virus an advantage, allowing it to escape the antibodies generated either by the disease or by the vaccine, without losing the ability to infect us efficiently. "When we looked at this data, there were two points that stood out as having the potential to harbor mutations that could be dangerous," adds the researcher. Next, the mutations were tested on the lab bench, where they were proven to have the potential to evade antibodies. "This wouldn't have been possible without close collaboration between experimental and computational work, which is essential for us to get ahead of the virus," points out Cláudio Soares.
"The interaction between viruses and our immune system resembles a game of rope, in which each of the "teams" pulls the rope in its direction. What we saw in this study is that certain mutations in the Spike protein of SARS-CoV-2 allow it to evade an important facet of the immune response, which is antibodies. However, the immune system is also plastic and adaptable. Knowing which mutations are most likely to escape the immune system allows us, if eventually necessary, to design strategies to help the immune system recognize and respond to these mutations more quickly."
One of the mutations noted, called 484, had been previously identified and is included in the variants of concern from Brazil, South Africa and India. The other mutation, 494, emerges as a new structural point likely to change in the SARS-CoV-2 virus. "Mutation 494 is on the list of variants currently under investigation by the CDC [Centers for Disease Control and Prevention] and Public Health England. With this study, we were able to prove that it allows the virus to escape antibodies. This proves that we should investigate the need to develop vaccines and therapies that can respond to these mutations, as well as determine the mechanisms that allow the virus to replicate without being recognized," adds Maria João Amorim.
The study was developed at the Instituto Gulbenkian de Ciência in collaboration with ITQB NOVA and the Chronic Diseases Research Centre - CEDOC. Funding was provided through grants PTDC/CCI-BIO/28200/2017 and FCT RESEARCH4COVID 19 (Ref 580), from the Foundation for Science and Technology (FCT, Portugal) as well as by the Calouste Gulbenkian Foundation.
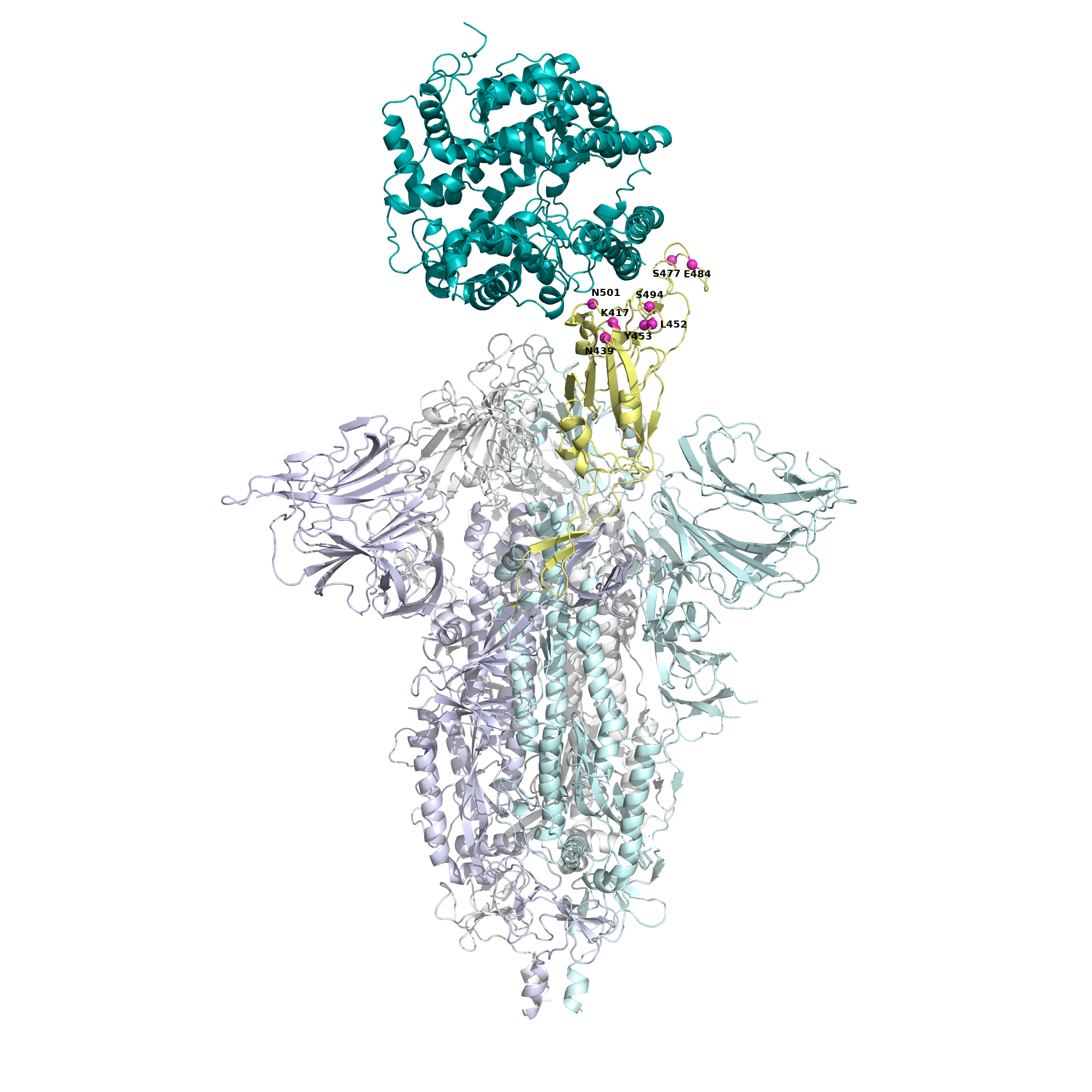
In the image the 3 "modules" of protein S in gray, lilac, and water green. The receptor-binding domain is in yellow with mutations marked in pink, and the ACE2 receptor, to which protein S binds, is colored "taupe.
Original Article:
PLOS Pathogens | 10.1101/2021.04.22.441007
Signatures in SARS-CoV-2 spike protein conferring escape to neutralizing antibodies
Marta Alenquer, Filipe Ferreira, Diana Lousa, Mariana Valério, Mónica Medina-Lopes, Marie-Louise Bergman, Juliana Gonçalves, Jocelyne Demengeot, Ricardo B. Leite, Jingtao Lilue, Zemin Ning, Carlos Penha-Gonçalves, Helena Soares, Cláudio M. Soares, Maria João Amorim







