Bacterial Cell Biology
The study of the subcellular organization of bacterial cells, over the last years, has revealed a surprising degree of protein compartmentalization and localization. Many essential cellular processes are performed by higher order protein complexes, which are precisely regulated in time and space. An example of such processes is cell division. For a cell to divide, it has to double its mass, replicate and segregate its genome and synthesize the division septum for the separation of the two daughter cells. Our laboratory uses mainly the pathogenic bacteria Staphylococcus aureus as a model for cell division and cell cycle studies, focusing on the processes of chromosome segregation and synthesis of the septum. For that purpose we use a range of genetic, biochemical and cell biology methods.
Of particular importance in our work is the use of super-resolution microscopy techniques. This is because S. aureus cells are very small, approximately 1micrometer in diameter. Therefore, many of the morphology changes that occur during the cell cycle of this bacterial pathogens, are beyond the resolution limit of conventional optical microscopy (~250nm). We use mainly Structured Illumination Microscopy (SIM), but also Photoactivated Localization Microscopy (PALM) to study live S. aureus cells.
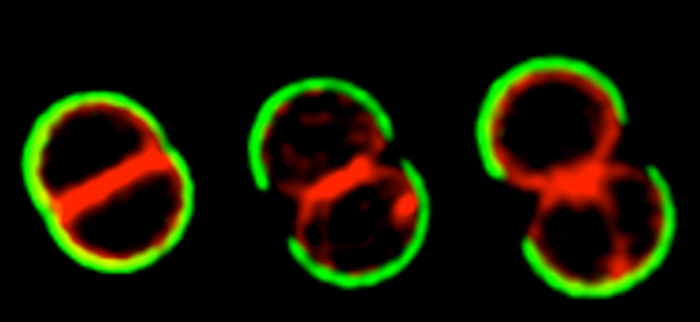
The cell cycle of Staphylococcus aureus
S. aureus COL cells were labeled with peripheral cell wall dye WGA-488 (green), washed, stained with membrane dye Nile Red and grown on the microscope slide. Cells were then imaged by Structured Illumination Microscopy (SIM)
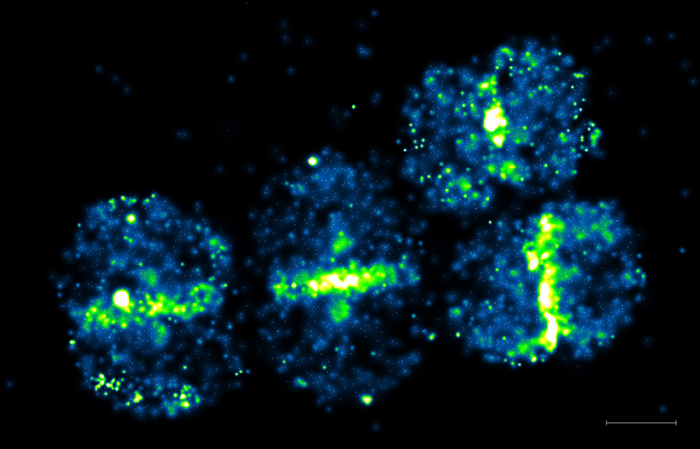
Single molecule detection of a protein involved in peptidoglycan synthesis
Penicillin Binding Protein 4 (PBP4) was tagged with Photoactivable mCherry and expressed Staphyloccus aureus. Live cells were observed by Photoactivated Localization Microscopy (PALM). Scale bar 500 nm.
Antibiotic resistance
Staphylococcus aureus is one of the most important community and hospital-acquired pathogens, with high morbidity and mortality rates being associated to staphylococcal infections. This problem is aggravated by the fact that many S. aureus clinical isolates have developed resistance against different classes of antibiotics. In fact, methicillin resistant S. aureus (MRSA) strains, which are resistant to beta-lactams and often to other classes of antibiotics, are on of the most important cause of antibiotic-resistant hospital infections worldwide.
Beta-lactams target the synthesis of the main component of the bacterial cell wall, peptidoglycan. The synthesis of peptidoglycan is the target of various classes of antibiotics, as it is an essential component virtually all bacteria and it is absent from eukaryotic cells. We want to understand the mechanism of action of existing and new antibacterial compounds, as well as the mechanisms of resistance developed by bacteria.
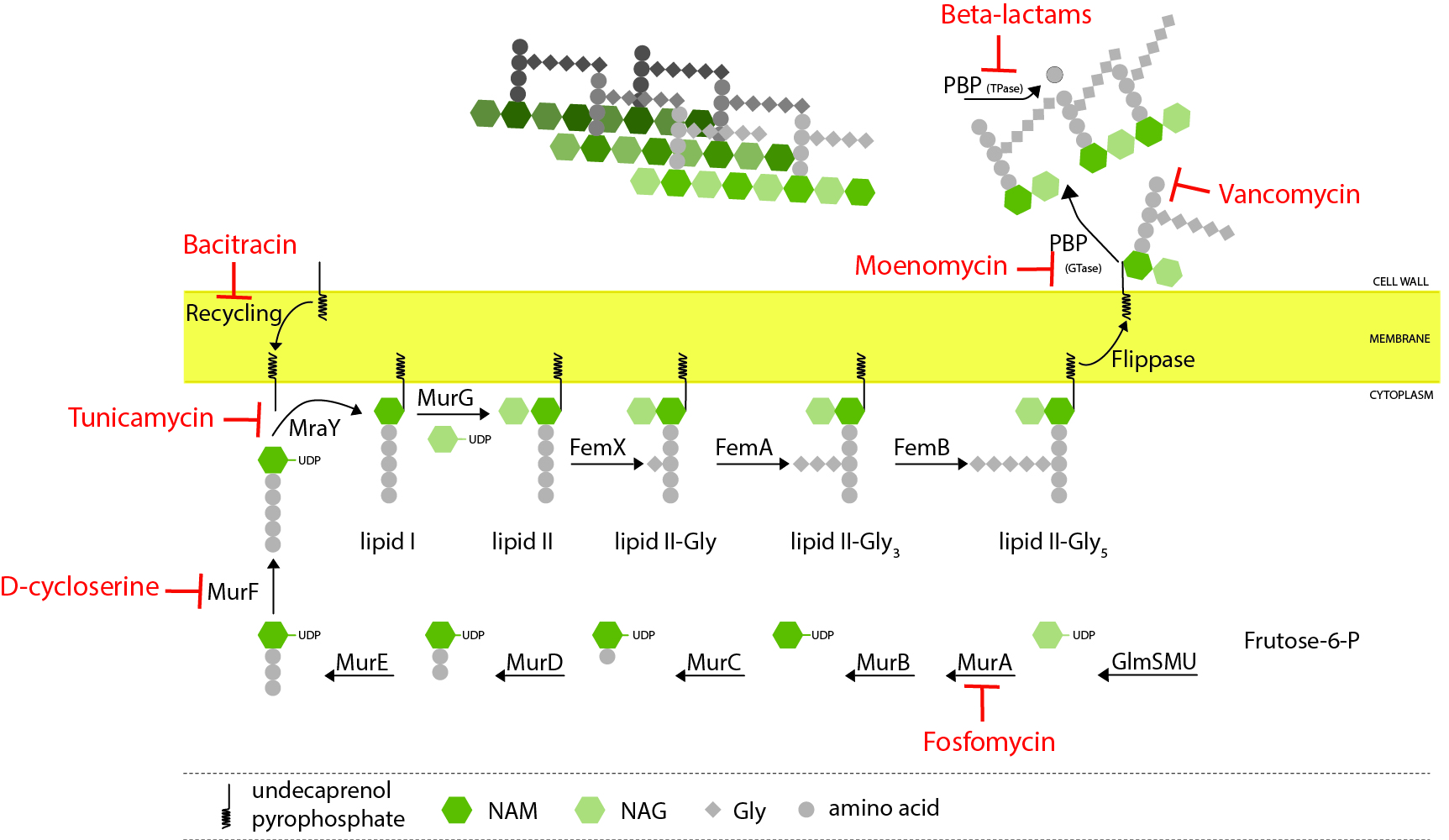
Peptidoglycan biosynthesis
Peptidoglycan biosynthesis starts in the cytoplasm with the formation of a soluble precursor which is then linked to the lipid carrier undecaprenol pyrophosphate. This lipid-linked precursor is flipped to the outside of the cell, where it is incorporated into the growing peptidoglycan, mainly by penicillin-binding proteins (PBPs). Various steps of this process can be inhibited by different classes of antibiotics (indicated in red)
For more detailed information on our science, please look at our Highlights.



