Superoxide Reductases (SORs)
Enzymes for Superoxide Detoxification in Anaerobes
First indication for SORs function was reported, in 1996, by Touati an co-workers, that while trying to isolate a SOD from the anaerobe D. baarsi , isolated a plasmid capable of protecting a sod deficient E.coli strain from the damage caused by superoxide (Pianzzola et al. 1996) . This plasmid contained the gene coding for a protein similar to desulfoferrodoxin (Dfx), first isolated from D. desulfuricans strain 27774 and from D. vulgaris Hildenborough (Moura et al. 1990), suggesting a possible role for these protein in the detoxification of superoxide in anaerobes. D. gigas neelaredoxin (Nlr), a protein with homology to the C-terminal domain of Dfx, was the first to be reported to have a SOD activity (1200 U.mg-1 ) (Silva et al. 1999). Its overexpression suppresses the deleterious effects of the lack of sodA and sodB products in E.coli (Silva et al. 2001) .
The role of these proteins in the mechanism of detoxification of superoxide had an important new insight when it was proposed that P. furiosus Nlr was a superoxide reductase instead of superoxide dismutase (Jenney et al. 1999) . Pulse radiolysis studies on D. baarsi Dfx (Lombard et al. 2000) and A. fulgidus Nlr (Abreu et al. 2000; Abreu et al. 2001) showed that the reduction of superoxide by these enzymes is virtually diffusion-limited, with a second order rate constant of ~ 1×10^9 M-1 s-1 . These enzymes are thus called superoxide reductases (SORs), catalyzing the following reaction:
![]()
SORs were so far found in genomes anaerobic or microaerophilic organisms, and are widely spread among the prokaryotes: Archaea and Bacteria. Although some of these organisms have also in their genome genes that code for canonical SODs, others don't, and so they thrust only in SOR proteins to protect them against superoxide radical.
Structure
The active site structure of SOR consists of an unusual Fe 2+ center, with a square pyramidal geometry, coordinated by four hystidines, one cysteine and a glutamate ligand that is present only in the oxidized state.

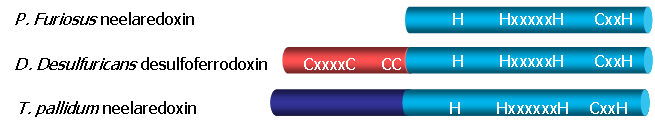
Three classes of SORs have been described, in accordance to the presence of an extra N-terminal domain that harbors another non-heme iron center. The homotetrameric Neelaredoxin ( Nlr ) family lacks this extra center, and the structure is already available for the protein from P.furiosus (Yeh, Hu et al. 2000). Desulfoferrodoxin ( Dfx ) family has a C-terminal domain homologous to Neelaredoxin, and a small N-terminal domain with a desulforedoxin-like iron center. This is a homodimeric protein and the structure was solved for D.desulfuricans (Coelho, Matias et al. 1997). The third family, isolated from the spirochete T. pallidum, contains the N-terminal domain, but lacks some of the cystein ligands for the second iron atom. The function of the extra center in Dfx is still unkown, as it appears that it doesn't contribute to the superoxide reduction mechanism.
SUPEROXIDE DETOXIFICATION IN Archaeoglobus fulgidus
Archaeoglobus members are hyperthermophiles that can be found in hydrothermal vents, oil deposits, and hot springs. They can produce biofilm when subjected to environmental stresses such as extreme pH or temperature, high concentrations of metal, or the addition of antibiotics, xenobiotics, or oxygen. These archaeons are known to cause the corrosion of iron and steel in oil and gas processing systems by producing iron sulphide. Their bioflims, however, may have industrial or research applications in the form of detoxifying metal contaminated samples or to gather metals in an economically recoverable form.
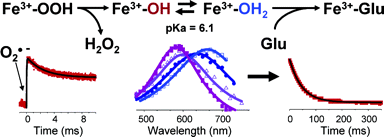
Analysis of the complete genome of A. fulgidus, reveals the presence of two types of SOR codifying genes: Dfx and Nlr. These proteins are being studied in our lab to understand the mechanism of superoxide reduction. By using pulse radiolysis and stopped-flow kinetics we were able to characterize some steps of the mechanism of superoxide reduction by SOR.
Click for more information on:
| Oxidative Stress and Anaerobes |
| Superoxide reductases |
PUBlCATIONS:
Rodrigues JV, Saraiva LM, Abreu IA, Teixeira M, Cabelli DE. Superoxide reduction by Archaeoglobus fulgidus desulfoferrodoxin: comparison with neelaredoxin. (2006) J Biol Inorg Chem in press
Rodrigues JV, Abreu IA, Cabelli D, Teixeira M. (2006). “Superoxide reduction mechanism of Archaeoglobus fulgidus one-iron superoxide reductase.” Biochemistry. 2006 Aug 1;45(30):9266-78
Rodrigues, J. V., Abreu, I. A., Saraiva, L. M., and Teixeira, M. (2005) Rubredoxin acts as an electron donor for neelaredoxin in Archaeoglobus fulgidus Biochem Biophys Res Commun 329 , 1300-5.
Abreu, I. A., L. M. Saraiva, et al. (2001). "The mechanism of superoxide scavenging by Archaeoglobus fulgidus neelaredoxin." J Biol Chem 276(42): 38995-9001.
Silva, G., LeGall, J., Xavier, A. V., Teixeira, M., and Rodrigues-Pousada, C. (2001) Molecular characterization of Desulfovibrio gigas neelaredoxin, a protein involved in oxygen detoxification in anaerobes J Bacteriol 183 , 4413-20.
Abreu, I. A., L. M. Saraiva, et al. (2000). "Oxygen detoxification in the strict anaerobic archaeon Archaeoglobus fulgidus : superoxide scavenging by neelaredoxin." Mol Microbiol 38(2): 322-34.
Silva, G., Oliveira, S., Gomes, C. M., Pacheco, I., Liu, M. Y., Xavier, A. V., Teixeira, M., Legall, J., and Rodrigues-pousada, C. (1999) Desulfovibrio gigas neelaredoxin. A novel superoxide dismutase integrated in a putative oxygen sensory operon of an anaerobe Eur J Biochem 259 , 235-43.
Coelho, A. V., P. Matias, et al. (1997). "Dfx structure determined by MAD phasing and refinement to 1.9-angstrom resolution reveals a unique combination of a tetrahedral FeS4 centre with a square pyramidal FeSN4 centre." J Biol Inorg Chem 2 : 680-689.
OTHER REFERENCES :
Niviere, V., Asso, M., Weill, C. O., Lombard, M., Guigliarelli, B., Favaudon, V., and Houee-Levin, C. (2004) Superoxide reductase from Desulfoarculus baarsii: identification of protonation steps in the enzymatic mechanism Biochemistry 43 , 808-18.
Emerson, J. P., Coulter, E. D., Cabelli, D. E., Phillips, R. S., and Kurtz, D. M., Jr. (2002) Kinetics and mechanism of superoxide reduction by two-iron superoxide reductase from Desulfovibrio vulgaris Biochemistry 41 , 4348-57.
Lombard, M., Fontecave, M., Touati, D., and Niviere, V. (2000) Reaction of the desulfoferrodoxin from Desulfoarculus baarsii with superoxide anion. Evidence for a superoxide reductase activity J Biol Chem 275 , 115-21.
Yeh, A. P., Hu, Y., Jenney, F. E., Jr., Adams, M. W., and Rees, D. C. (2000) Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states, Biochemistry 39, 2499-508.
Jenney, F. E., Jr., Verhagen, M. F., Cui, X., and Adams, M. W. (1999) Anaerobic microbes: oxygen detoxification without superoxide dismutase Science 286 , 306-9.
Klenk HP , Clayton RA , Tomb JF , White O , Nelson KE , Ketchum KA , Dodson RJ , Gwinn M , Hickey EK , Peterson JD , Richardson DL , Kerlavage AR , Graham DE , Kyrpides NC , Fleischmann RD , Quackenbush J , Lee NH , Sutton GG , Gill S , Kirkness EF , Dougherty BA , McKenney K , Adams MD , Loftus B , Peterson S , Reich CI , McNeil LK , Badger JH , Glodek A , Zhou L , Overbeek R , Gocayne JD , Weidman JF , McDonald L , Utterback T , Cotton MD , Spriggs T , Artiach P , Kaine BP , Sykes SM , Sadow PW , D'Andrea KP , Bowman C , Fujii C , Garland SA , Mason TM , Olsen GJ , Fraser CM , Smith HO , Woese CR , Venter JC . (1997) The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390(6658):364-70
Pianzzola, M. J., Soubes, M., and Touati, D. (1996) Overproduction of the rbo gene product from Desulfovibrio species suppresses all deleterious effects of lack of superoxide dismutase in Escherichia coli J Bacteriol 178 , 6736-42.
Moura, I., Tavares, P., Moura, J. J., Ravi, N., Huynh, B. H., Liu, M. Y., and LeGall, J. (1990) Purification and characterization of desulfoferrodoxin. A novel protein from Desulfovibrio desulfuricans (ATCC 27774) and from Desulfovibrio vulgaris (strain Hildenborough) that contains a distorted rubredoxin center and a mononuclear ferrous center J Biol Chem 265 , 21596-602
MicrobeWiki: http://microbewiki.kenyon.edu/index.php/MicrobeWiki