3D structure of anti-tuberculosis target makes cover of Molecular Cell
According to the World Health Organization, in 2018 alone an estimated 10 million people were infected with Tuberculosis (TB) worldwide, making this disease one of the top 10 causes of death caused by a single infectious agent, the bacteria Mycobacterium tuberculosis. Its cell envelope is crucial for growth and virulence of M. tuberculosis and is also a major contributor to resistance to common antibiotics. The importance of this unique structure in mycobacteria makes its biosynthetic enzymes attractive drug targets.

Archer Lab from the Macromolecular Crystallography Unit at ITQB NOVA is involved in a consortium, led by the Mancia group from Columbia University and New York Structural Biology Center (USA), which aims to unravel the structure of Arabinofuranosyltransferases, together with researchers from Simons Electron Microscopy Center (USA), iMed.ULisboa (Portugal), Alberta University (Canada) and University of Alabama at Birmingham (USA).
The structural and functional characterization of AftD has been published in Molecular Cell. An artistic image of AftD, created by a former ITQB NOVA PhD student, has made cover of this prestigious journal in Life Sciences.
This characterization was possible using Cryo-electron microscopy (cryo-EM). Recent advances in this technology has allowed the determination of biomolecular structures at near-atomic resolution. Through single-particle cryo-EM method, images from several thin sections from the AftD protein were collected and processed into a combined representative image, using a specific computer algorithm. The overall AftD protein architecture at 2.9 Angstrom (Å) enabled the identification of important functional sites and revealed an unexpected tight association with an acyl carrier protein (ACP), which seems to act as a functional regulator of this enzyme.
As AftD is an important enzyme for the latest step of the mycobacteria envelop biosynthesis and some point mutations on its gene may confer drug resistance to M. tuberculosis, this consortium set up to better understand the mechanisms of resistance as well as to design new drugs targeting this enzyme. “These results are very promising. The identification of the putative active sites and ACP binding residues open new perspectives to design specific drugs and control the tuberculosis infection”, explains Margarida Archer. Besides tuberculosis, this study may also be relevant for diseases such as diphtheria and leprosy, caused by similar pathogenic bacteria.
The collaboration with Mancia group continues and also includes training students on the emerging cryo-EM technology supported by PROMETEUS, an EU H2020 RISE project. At ITQB NOVA, work is underway within an FCT project coordinated by M. Archer and also with the support of EU H2020 Instruct-Ultra project, in collaboration with Cláudio M. Soares group (in silico computational studies) and Rita Ventura group (chemical synthesis).
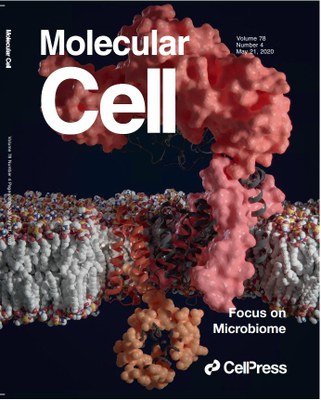
Cover of Molecular Cell n.4 May 21, 2020 - 3D model of membrane enzyme AftD embedded in the membrane of mycobacterium, solved by cryogenic electron microscopy. This protein is essential for building the bacterial cell wall and has a large periplasmic domain (pink upper surface) to bind to complex glycans. The catalytic core of the enzyme is closer to the eleven transmembrane helices which has a conserved glycosyltransferase fold (gray). Unexpectedly, this enzyme is regulated by another soluble protein, the acyl carrier protein (orange dots), which interact with AftD C-terminal helices (darker orange). Artwork by Davide Cruz
In the news
- Desvendada estrutura de proteína essencial na bactéria da tuberculose, in Público 25.05.2020
Original paper
Yong Zi Tan, Lei Zhang, José Rodrigues, Ruixiang Blake Zheng, Sabrina I. Giacometti, Ana L. Rosário, Brian Klos, Venkata P. Dandey, Hui Wei, Richard Brunton, Ashleigh M. Raczkowski, Diogo Athayde, Maria João Catalão, Madalena Pimentel, Oliver B. Clarke, Tod L. Lowary , Margarida Archer , Michael Niederweis , Clinton S. Potter, Bridget Carragher, Filippo Mancia
Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria
Molecular Cell doi: 10.1016/j.molcel.2020.04.014







